We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
This is a default text for your page '''Isabelle A. Altieri/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Isabelle A. Altieri/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | ==PDB structure== | ||
| - | <scene name='78/781947/Biological_assembly_1/1'>biological assembly 1</scene> | ||
| - | |||
| - | <scene name='78/781947/Hydrophilic_residues/2'>Asp70 Asp111 Asp117</scene> | ||
| - | |||
| - | <scene name='78/781947/H1_and_h2_h2ophobic_residues/2'>Phe 74 Phe 122 Met 158 Met 161 Leu163</scene> | ||
| - | |||
| - | <scene name='78/781947/Pro-ser_in_linker/1'>Pro91-Ser92</scene>site where eIF4 targets for phosphorylation when interacting in initiation translation | ||
| - | <scene name='78/781947/Residues_interacting_with_a6/1'>interactions with A6</scene> | ||
| - | <scene name='78/781947/Interactions_with_a3/1'>interactions with A3</scene> | ||
| - | |||
==Introduction== | ==Introduction== | ||
| Line 30: | Line 19: | ||
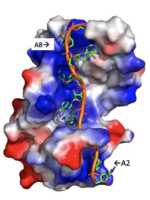
A primary function of PABP is recognizing and interacting with the 3'poly (A) tail created in mRNA processing. As found by EMSA competition experiments, there is a minimum of 11-12 adenosines necessary in the poly (A) tail for the adenosine chain to bind to PABP with high affinity. However, for one biological assembly, a chain containing 9 adenosines sufficiently binds the assembly for crystallization and is shown in the biological assembly structure. The 4 RRM domains that are the primary interacting sites for the adenosine recognition exist as globular domains, each having four antiparallel β-strands and two α-helices. With the N-terminal to C-terminal motifs labeled as S1 to S4 for the β-strands and H1 to H2 for the α-helices, the strands are spatially arranged as S2-S3-S1-S4. Furthermore, there are two conserved sequences in each RRM, called RNP1 and 2. RNP 1 consists of a conserved sequence of 8 residues, while RNP2 consists of a conserved sequence of 6 residues. Much of the weak intermolecular interactions with adenosine from the RRMs occur from the <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2</scene> conserved sequences, which correspond to the two central β-strands, with specific interactions shown in Figure 2.The support for adenosine recognition by the RRMs occurs as a type of binding trough with the sheets, primarily <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2 forming the Primary Binding Trough</scene>, and the interstrand loop between β-strands 2 and 3 as well as the domain linker forming the <scene name='78/781946/Adenosine_binding_wall/1'> Adenosine Binding Wall</scene>. Additionally, the primary binding trough is stabilized by <scene name='78/781946/Rrm1_2_packing_intxn/2'>Stabilizing Packing Interactions of RRM1 RRM2 Binding Trough</scene>. | A primary function of PABP is recognizing and interacting with the 3'poly (A) tail created in mRNA processing. As found by EMSA competition experiments, there is a minimum of 11-12 adenosines necessary in the poly (A) tail for the adenosine chain to bind to PABP with high affinity. However, for one biological assembly, a chain containing 9 adenosines sufficiently binds the assembly for crystallization and is shown in the biological assembly structure. The 4 RRM domains that are the primary interacting sites for the adenosine recognition exist as globular domains, each having four antiparallel β-strands and two α-helices. With the N-terminal to C-terminal motifs labeled as S1 to S4 for the β-strands and H1 to H2 for the α-helices, the strands are spatially arranged as S2-S3-S1-S4. Furthermore, there are two conserved sequences in each RRM, called RNP1 and 2. RNP 1 consists of a conserved sequence of 8 residues, while RNP2 consists of a conserved sequence of 6 residues. Much of the weak intermolecular interactions with adenosine from the RRMs occur from the <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2</scene> conserved sequences, which correspond to the two central β-strands, with specific interactions shown in Figure 2.The support for adenosine recognition by the RRMs occurs as a type of binding trough with the sheets, primarily <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2 forming the Primary Binding Trough</scene>, and the interstrand loop between β-strands 2 and 3 as well as the domain linker forming the <scene name='78/781946/Adenosine_binding_wall/1'> Adenosine Binding Wall</scene>. Additionally, the primary binding trough is stabilized by <scene name='78/781946/Rrm1_2_packing_intxn/2'>Stabilizing Packing Interactions of RRM1 RRM2 Binding Trough</scene>. | ||
| - | |||
[[Image: Adenosine_backbone.png |150 px|right|thumb|Figure 3: Basic residues of RRM 1 and 2 make stabilizing electrostatic interactions with the negatively charged adenosine phosphates. ]] | [[Image: Adenosine_backbone.png |150 px|right|thumb|Figure 3: Basic residues of RRM 1 and 2 make stabilizing electrostatic interactions with the negatively charged adenosine phosphates. ]] | ||
| + | ====Adenosine Stabilization Interaction Patterns==== | ||
| + | |||
| + | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between aromatic and alipathic side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by Lysine 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88. | ||
| + | |||
| + | ===Translation Initiation=== | ||
| + | <scene name='78/781946/Pabp_linker_conserved_residues/1'>PABP linker with Conserved Residues Shown</scene> | ||
| + | |||
| + | <scene name='78/781947/H1_and_h2_h2ophobic_residues/2'>Phe 74 Phe 122 Met 158 Met 161 Leu163</scene> | ||
| + | <scene name='78/781947/Hydrophilic_residues/2'>Asp70 Asp111 Asp117</scene> | ||
== Disease == | == Disease == | ||
| Line 38: | Line 35: | ||
== Relevance == | == Relevance == | ||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | |||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 03:57, 29 March 2018
Poly(A) binding protein
Structure
| |||||||||||