We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
====Adenosine Stabilization Interaction Patterns==== | ====Adenosine Stabilization Interaction Patterns==== | ||
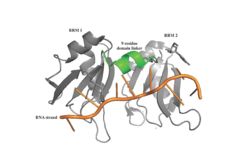
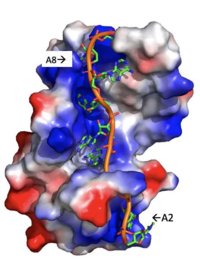
| - | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between aromatic and alipathic side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by Lysine 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88. <ref> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2. <ref/> | + | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between aromatic and alipathic side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by Lysine 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88. <ref> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2. <ref/>. |
===Translation Initiation=== | ===Translation Initiation=== | ||
| Line 39: | Line 39: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | |||
| - | 1. Blatter, Markus, et al. “RNA Recognition Motifs: Boring? Not Quite.” Current Opinion in Structural Biology, vol. 18, no. 3, 2008, pp. 290–298., doi:10.1016/j.sbi.2008.04.002. | ||
| - | |||
| - | 2.De Melo Neto, Osvaldo P., et al. “Phosphorylation and Interactions Associated with the Control of the Leishmania Poly-A Binding Protein 1 (PABP1) Function during Translation Initiation.” RNA Biology, 23 Mar. 2018, pp. 1–17., doi:10.1080/15476286.2018.1445958. | ||
| - | |||
| - | 3.Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2. | ||
| - | |||
| - | 4.Gallie, Daniel R, and Renyi Liu. “Phylogenetic Analysis Reveals Dynamic Evolution of the Poly(A)-Binding Protein Gene Family in Plants.” BMC Evolutionary Biology, vol. 14, no. 1, 2014, doi:10.1186/s12862-014-0238-4. | ||
| - | |||
| - | 5.Gingras, Anne-Claude, et al. “eIF4 Initiation Factors: Effectors of MRNA Recruitment to Ribosomes and Regulators of Translation.” Annual Review of Biochemistry, vol. 68, no. 1, 1999, pp. 913–963., doi:10.1146/annurev.biochem.68.1.913. | ||
| - | |||
| - | 6.Imataka, H. “A Newly Identified N-Terminal Amino Acid Sequence of Human eIF4G Binds Poly(A)-Binding Protein and Functions in Poly(A)-Dependent Translation.” The EMBO Journal, vol. 17, no. 24, 1998, pp. 7480–7489., doi:10.1093/emboj/17.24.7480. | ||
| - | |||
| - | 7.Kahvejian, A. “Mammalian Poly(A)-Binding Protein Is a Eukaryotic Translation Initiation Factor, Which Acts via Multiple Mechanisms.” Genes & Development, vol. 19, no. 1, 2005, pp. 104–113., doi:10.1101/gad.1262905. | ||
| - | |||
| - | 8. Piron, M. “Rotavirus RNA-Binding Protein NSP3 Interacts with eIF4GI and Evicts the Poly(A) Binding Protein from eIF4F.” The EMBO Journal, vol. 17, no. 19, 1998, pp. 5811–5821., doi:10.1093/emboj/17.19.5811. | ||
Revision as of 17:32, 29 March 2018
Poly(A) binding protein - Homo sapiens
Structure
| |||||||||||