We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
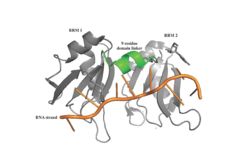
[[Image: PABP Biological Assembly 1.jpg |250 px|left|thumb|Figure 1: PABP Biological Assembly with linker highlighted. ]] | [[Image: PABP Biological Assembly 1.jpg |250 px|left|thumb|Figure 1: PABP Biological Assembly with linker highlighted. ]] | ||
| - | Human Poly(A) Binding Protein (PABP) is a biopolypeptide involved in recognizing the 3'poly (A) tail of mRNA that is added to an mRNA transcript during mRNA processing.This recognition as well as PABP's interaction with other proteins and initiation factors causes it to also play a significant role in translation initiation and mRNA stabilization and degradation. PABP consists of four conserved domains of RNA recognition motifs (RRMs); however, the two N-terminal RRMs (RRM1 and RRM2) and the short linker sequence that connects them supports most of the function of PABP, so RRM3 and RRM4 may not be essential. Thus, the published X-ray structure exhibits RRM1 and RRM2 at a 2.6 Angstrom resolution. This is shown as <scene name='78/781947/Biological_assembly_1/1'>Biological Assembly 1</scene>. Both RRM 1 and 2 are needed to support biochemical function, that is, no one RRM can support biochemical function. Additionally, there is a proline rich C-terminal portion of variable length that is not well conserved and unknown as to how it contributes to the protein's function <ref name= "PABP"> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2.</ref>. | + | Human Poly(A) Binding Protein (PABP) is a biopolypeptide involved in recognizing the [https://en.wikipedia.org/wiki/Polyadenylation 3' poly(A) tail] of [https://en.wikipedia.org/wiki/Messenger_RNA mRNA] that is added to an mRNA transcript during mRNA processing.This recognition as well as PABP's interaction with other proteins and [https://en.wikipedia.org/wiki/Initiation_factor initiation factors] causes it to also play a significant role in [https://en.wikipedia.org/wiki/Eukaryotic_translation translation] initiation and mRNA stabilization and degradation. PABP consists of four conserved [https://en.wikipedia.org/wiki/Protein_domain domains] of [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motifs] (RRMs); however, the two [https://en.wikipedia.org/wiki/N-terminus N-terminal] RRMs (RRM1 and RRM2) and the short linker sequence that connects them supports most of the function of PABP, so RRM3 and RRM4 may not be essential. Thus, the published X-ray structure, found by [https://en.wikipedia.org/wiki/X-ray_scattering_techniques X-ray diffraction], exhibits RRM1 and RRM2 at a 2.6 Angstrom [https://en.wikipedia.org/wiki/Resolution_(electron_density) resolution]. This is shown as <scene name='78/781947/Biological_assembly_1/1'>Biological Assembly 1</scene>. Both RRM 1 and 2 are needed to support biochemical function, that is, no one RRM can support biochemical function. Additionally, there is a [https://en.wikipedia.org/wiki/Proline proline] rich [https://en.wikipedia.org/wiki/C-terminus C-terminal] portion of variable length that is not well conserved and unknown as to how it contributes to the protein's function <ref name= "PABP"> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2.</ref>. |
== Function == | == Function == | ||
| Line 18: | Line 18: | ||
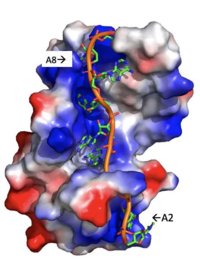
[[Image: Stacking_of_adenosines_with_beta_sheets.jpg |250 px|left|thumb|Figure 2: The specific weak intermolecular interactions between RNP1 and RNP2 and Adenosines. These interactions are the primary support of adenosine recognition by PABP and include mainly van der Waals interactions, hydrogen bonds, and stacking interactions. ]] | [[Image: Stacking_of_adenosines_with_beta_sheets.jpg |250 px|left|thumb|Figure 2: The specific weak intermolecular interactions between RNP1 and RNP2 and Adenosines. These interactions are the primary support of adenosine recognition by PABP and include mainly van der Waals interactions, hydrogen bonds, and stacking interactions. ]] | ||
| - | A primary function of PABP is recognizing and interacting with the 3'poly (A) tail created in mRNA processing. As found by EMSA competition experiments, there is a minimum of 11-12 adenosines necessary in the poly (A) tail for the adenosine chain to bind to PABP with high affinity. However, for one biological assembly, a chain containing 9 adenosines sufficiently binds the assembly for crystallization and is shown in the biological assembly structure. The 4 RRM domains that are the primary interacting sites for the adenosine recognition exist as globular domains, each having four antiparallel β-strands and two α-helices. With the N-terminal to C-terminal motifs labeled as S1 to S4 for the β-strands and H1 to H2 for the α-helices, the strands are spatially arranged as S2-S3-S1-S4. Furthermore, there are two conserved sequences in each RRM, called RNP1 and 2. RNP 1 consists of a conserved sequence of 8 residues, while RNP2 consists of a conserved sequence of 6 residues. Much of the weak intermolecular interactions with adenosine from the RRMs occur from the <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2</scene> conserved sequences, which correspond to the two central β-strands, with specific interactions shown in Figure 2.The support for adenosine recognition by the RRMs occurs as a type of binding trough with the sheets, primarily <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2 forming the Primary Binding Trough</scene>, and the interstrand loop between β-strands 2 and 3 as well as the domain linker forming the <scene name='78/781946/Adenosine_binding_wall/1'> Adenosine Binding Wall</scene>. Additionally, the primary binding trough is stabilized by <scene name='78/781946/Rrm1_2_packing_intxn/2'>Stabilizing Packing Interactions of RRM1 RRM2 Binding Trough</scene>. <ref name= "PABP"/> | + | A primary function of PABP is recognizing and interacting with the 3'poly (A) tail created in mRNA processing. As found by [https://en.wikipedia.org/wiki/Electrophoretic_mobility_shift_assay EMSA competition experiments], there is a minimum of 11-12 adenosines necessary in the poly (A) tail for the adenosine chain to bind to PABP with high affinity. However, for one biological assembly, a chain containing 9 adenosines sufficiently binds the assembly for [https://en.wikipedia.org/wiki/Crystallization crystallization] and is shown in the biological assembly structure. The 4 RRM domains that are the primary interacting sites for the adenosine recognition exist as globular domains, each having four antiparallel [https://en.wikipedia.org/wiki/Beta_sheet β-strands] and two [https://en.wikipedia.org/wiki/Alpha_helix α-helices]. With the N-terminal to C-terminal motifs labeled as S1 to S4 for the β-strands and H1 to H2 for the α-helices, the strands are spatially arranged as S2-S3-S1-S4. Furthermore, there are two [https://en.wikipedia.org/wiki/Conserved_sequence conserved sequences] in each RRM, called RNP1 and 2. RNP 1 consists of a conserved sequence of 8 [https://en.wikipedia.org/wiki/Residue residues], while RNP2 consists of a conserved sequence of 6 residues. Much of the weak [https://en.wikipedia.org/wiki/Intermolecular_force intermolecular interactions] with [https://en.wikipedia.org/wiki/Adenosine adenosine] from the RRMs occur from the <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2</scene> conserved sequences, which correspond to the two central β-strands, with specific interactions shown in Figure 2.The support for adenosine recognition by the RRMs occurs as a type of binding trough with the sheets, primarily <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2 forming the Primary Binding Trough</scene>, and the interstrand loop between β-strands 2 and 3 as well as the domain linker forming the <scene name='78/781946/Adenosine_binding_wall/1'> Adenosine Binding Wall</scene>. Additionally, the primary binding trough is stabilized by <scene name='78/781946/Rrm1_2_packing_intxn/2'>Stabilizing Packing Interactions of RRM1 RRM2 Binding Trough</scene>. <ref name= "PABP"/> |
[[Image: Adenosine_backbone.png |200 px|right|thumb|Figure 3: Basic residues of RRM 1 and 2 (shown in blue) make stabilizing electrostatic interactions with the negatively charged adenosine phosphates. ]] | [[Image: Adenosine_backbone.png |200 px|right|thumb|Figure 3: Basic residues of RRM 1 and 2 (shown in blue) make stabilizing electrostatic interactions with the negatively charged adenosine phosphates. ]] | ||
====Adenosine Stabilization Interaction Patterns==== | ====Adenosine Stabilization Interaction Patterns==== | ||
| - | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between aromatic and alipathic side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by Lysine 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88 <ref name= "PABP"/>. | + | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between [https://en.wikipedia.org/wiki/Aromatic_amino_acid aromatic] and [https://en.wikipedia.org/wiki/Aliphatic_compound alipathic] side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by [https://en.wikipedia.org/wiki/Lysine Lysine] 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88 <ref name= "PABP"/>. |
===Translation Initiation=== | ===Translation Initiation=== | ||
| Line 39: | Line 39: | ||
== Medical Relevancy == | == Medical Relevancy == | ||
==='''Rotavirus' Effect on Initiation of Translation'''=== | ==='''Rotavirus' Effect on Initiation of Translation'''=== | ||
| - | The initiation of translation in eukaryotes is supported by a closed loop model. This model requires the 5' end and the 3' end of mRNA to be physically connected. The poly(A)-binding protein is necessary for initiation of translation and is required for the closed loop model. Rotavirus, a virus of varying size, containing 11 double stranded RNA and 12 proteins (6 structural, 6 non-structural) is responsible for preventing initiation of translation in infected cells. The virus enters the cell and undergoes a non-conservative replication cycle in the cytoplasm. After a replication cycle non-structural protein 3 (NSP3) can be found spread throughout the cytoplasm. NSP3 is responsible for releasing PABP from eIF4F and inhibiting translation initiation. In a study done by Piron et al. it has been seen that NSP3 competes with PABP in binding to the poly(A)-tail of mRNA. This competitor inhibits the proper closing of the closed loop therefore inhibiting translation and protein synthesis. Not only does the rotavirus inhibit protein synthesis of the host cell but it successfully initiatives its own translation as well. The viral mRNA and the host translation initiation factors are in close enough proximity to allow the viral mRNA bound to NSP3 to undergo translation. The translation of viral mRNA allows the virus to spread throughout an organism and lead to a greater decrease in host protein synthesis. When infected with rotavirus one may experience diarrhea, fever, vomiting, and dehydration. Without an antiviral it is suggested to increase fluid intake and allow three to seven days for the infection to subside. <ref name="Rotavirus"> Piron, M. “Rotavirus RNA-Binding Protein NSP3 Interacts with eIF4GI and Evicts the Poly(A) Binding Protein from eIF4F.” The EMBO Journal, vol. 17, no. 19, 1998, pp. 5811–5821., doi:10.1093/emboj/17.19.5811. </ref> | + | The initiation of translation in [https://en.wikipedia.org/wiki/Eukaryote eukaryotes] is supported by a closed loop model. This model requires the 5' end and the 3' end of mRNA to be physically connected. The poly(A)-binding protein is necessary for initiation of translation and is required for the closed loop model. [https://en.wikipedia.org/wiki/Rotavirus Rotavirus], a [https://en.wikipedia.org/wiki/Virus virus] of varying size, containing 11 double stranded RNA and 12 proteins (6 structural, 6 non-structural) is responsible for preventing initiation of translation in infected cells. The virus enters the cell and undergoes a non-conservative [https://en.wikipedia.org/wiki/Viral_replication replication cycle] in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm]. After a replication cycle non-structural protein 3 (NSP3) can be found spread throughout the cytoplasm. NSP3 is responsible for releasing PABP from eIF4F and inhibiting translation initiation. In a study done by Piron et al. it has been seen that NSP3 competes with PABP in binding to the poly(A)-tail of mRNA. This competitor inhibits the proper closing of the closed loop therefore inhibiting translation and [https://en.wikipedia.org/wiki/Protein_biosynthesis protein synthesis]. Not only does the rotavirus inhibit protein synthesis of the host cell but it successfully initiatives its own translation as well. The viral mRNA and the host translation initiation factors are in close enough proximity to allow the viral mRNA bound to NSP3 to undergo translation. The translation of viral mRNA allows the virus to spread throughout an [https://en.wikipedia.org/wiki/Organism organism] and lead to a greater decrease in host protein synthesis. When infected with rotavirus one may experience [https://en.wikipedia.org/wiki/Diarrhea diarrhea], [https://en.wikipedia.org/wiki/Fever fever], [https://en.wikipedia.org/wiki/Vomiting vomiting], and [https://en.wikipedia.org/wiki/Dehydration dehydration]. Without an [https://en.wikipedia.org/wiki/Antiviral_drug antiviral] it is suggested to increase fluid intake and allow three to seven days for the infection to subside. <ref name="Rotavirus"> Piron, M. “Rotavirus RNA-Binding Protein NSP3 Interacts with eIF4GI and Evicts the Poly(A) Binding Protein from eIF4F.” The EMBO Journal, vol. 17, no. 19, 1998, pp. 5811–5821., doi:10.1093/emboj/17.19.5811. </ref> |
== Biological Relevancy == | == Biological Relevancy == | ||
==='''Poly(A) Binding Protein's Evolution in plants'''=== | ==='''Poly(A) Binding Protein's Evolution in plants'''=== | ||
Revision as of 18:53, 29 March 2018
Poly(A) binding protein
Structure
| |||||||||||