We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 40: | Line 40: | ||
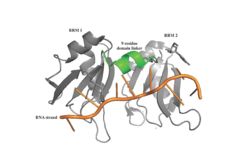
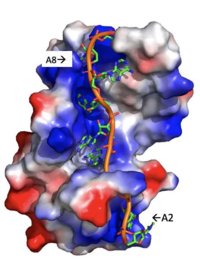
The RRMs support interactions with the interacting proteins such as eIF4G and [https://en.wikipedia.org/wiki/PAIP1 PAIP-1], however, the specific ways in which PABP interacts with these proteins are not structurally proven. However, there is a convex dorsal surface present on the RRM 1 and 2 motifs formed by the two α-helices in each RRM, specified as H1 and H2 in RRM1 and H1' and H2' in RRM2. This surface contains a sequence portion of <scene name='78/781947/H1_and_h2_h2ophobic_residues/2'>Conserved Hydrophobic Residues</scene>and <scene name='78/781947/Hydrophilic_residues/2'>Conserved Acidic Residues</scene> | The RRMs support interactions with the interacting proteins such as eIF4G and [https://en.wikipedia.org/wiki/PAIP1 PAIP-1], however, the specific ways in which PABP interacts with these proteins are not structurally proven. However, there is a convex dorsal surface present on the RRM 1 and 2 motifs formed by the two α-helices in each RRM, specified as H1 and H2 in RRM1 and H1' and H2' in RRM2. This surface contains a sequence portion of <scene name='78/781947/H1_and_h2_h2ophobic_residues/2'>Conserved Hydrophobic Residues</scene>and <scene name='78/781947/Hydrophilic_residues/2'>Conserved Acidic Residues</scene> | ||
| - | residues. It is thought that this area of conservation thus produces overlapping binding sites to interact with eIF4G and PAIP-1. For example, the conserved acidic residues may be beneficial to be used in to interact with essential basic residues present in both eIF4G and PAIP-1 via ionic interactions <ref name="PABP"/>. | + | residues. It is thought that this area of conservation thus produces overlapping binding sites to interact with eIF4G and PAIP-1. For example, the conserved acidic residues may be beneficial to be used in to interact with essential basic residues present in both eIF4G and PAIP-1 via ionic interactions <ref name="PABP"/>. While there are only proposed mechanisms for how PABP promotes the initiation of translation in Homo sapiens, there is a pathogenic [https://en.wikipedia.org/wiki/Protozoa protozoan], Leishmania, that through a study done by Osvaldo P. de Melo Neto et. al, a site on the domain linker of a PABP homolog, PABP-1, of either serine-proline or threonine-proline residues which an eIF4F-like complex [https://en.wikipedia.org/wiki/Phosphorylation phosphorylates]. The authors suggest that this phosphorylation is part of how PABP-1 aids the eIF4F complex in initiating translation. They supported this by removing the gene that encodes PABP-1 and the results showed that the protozoan could not initiated cell growth and therefore survive without the PABP-1 gene. The homo sapiens' PABP also contain a serine-proline (<scene name='78/781947/Pro-ser_in_linker/2'>Pro91Ser92</scene>) site on it domain linker which could be interacting with the eIF4 complex in a similar way as in Leishmania protozoan. |
| Line 46: | Line 46: | ||
== Medical Relevancy == | == Medical Relevancy == | ||
==='''Rotavirus' Effect on Initiation of Translation'''=== | ==='''Rotavirus' Effect on Initiation of Translation'''=== | ||
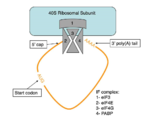
| - | The initiation of translation in [https://en.wikipedia.org/wiki/Eukaryote eukaryotes] is supported by a closed loop model. This model requires the 5' end and the 3' end of mRNA to be physically connected. The poly(A)-binding protein is necessary for initiation of translation and is required for the closed loop model. [https://en.wikipedia.org/wiki/Rotavirus Rotavirus], a [https://en.wikipedia.org/wiki/Virus virus] of varying size, containing 11 double stranded RNA and 12 proteins (6 structural, 6 non-structural) is responsible for preventing initiation of translation in infected cells. The virus enters the cell and undergoes a non-conservative [https://en.wikipedia.org/wiki/Viral_replication replication cycle] in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm]. After a replication cycle non-structural protein 3 (NSP3) can be found spread throughout the cytoplasm. NSP3 is responsible for releasing PABP from eIF4F and inhibiting translation initiation. In a study done by Piron et al. it has been seen that NSP3 competes with PABP in binding to the poly(A)-tail of mRNA. This competitor inhibits the proper closing of the closed loop therefore inhibiting translation and [https://en.wikipedia.org/wiki/Protein_biosynthesis protein synthesis]. Not only does the rotavirus inhibit protein synthesis of the host cell but it successfully initiatives its own translation as well. The viral mRNA and the host translation initiation factors are in close enough proximity to allow the viral mRNA bound to NSP3 to undergo translation. The translation of viral mRNA allows the virus to spread throughout an [https://en.wikipedia.org/wiki/Organism organism] and lead to a greater decrease in host protein synthesis. When infected with rotavirus one may experience | + | The initiation of translation in [https://en.wikipedia.org/wiki/Eukaryote eukaryotes] is supported by a closed loop model. This model requires the 5' end and the 3' end of mRNA to be physically connected. The poly(A)-binding protein is necessary for initiation of translation and is required for the closed loop model. [https://en.wikipedia.org/wiki/Rotavirus Rotavirus], a [https://en.wikipedia.org/wiki/Virus virus] of varying size, containing 11 double stranded RNA and 12 proteins (6 structural, 6 non-structural) is responsible for preventing initiation of translation in infected cells. The virus enters the cell and undergoes a non-conservative [https://en.wikipedia.org/wiki/Viral_replication replication cycle] in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm]. After a replication cycle non-structural protein 3 (NSP3) can be found spread throughout the cytoplasm. NSP3 is responsible for releasing PABP from eIF4F and inhibiting translation initiation. In a study done by Piron et al. it has been seen that NSP3 competes with PABP in binding to the poly(A)-tail of mRNA. This competitor inhibits the proper closing of the closed loop therefore inhibiting translation and [https://en.wikipedia.org/wiki/Protein_biosynthesis protein synthesis]. Not only does the rotavirus inhibit protein synthesis of the host cell but it successfully initiatives its own translation as well. The viral mRNA and the host translation initiation factors are in close enough proximity to allow the viral mRNA bound to NSP3 to undergo translation. The translation of viral mRNA allows the virus to spread throughout an [https://en.wikipedia.org/wiki/Organism organism] and lead to a greater decrease in host protein synthesis. When infected with rotavirus one may experience diarrhea,fever, vomiting, and dehydration. Without an [https://en.wikipedia.org/wiki/Antiviral_drug antiviral] it is suggested to increase fluid intake and allow three to seven days for the infection to subside. <ref name="Rotavirus"> Piron, M. “Rotavirus RNA-Binding Protein NSP3 Interacts with eIF4GI and Evicts the Poly(A) Binding Protein from eIF4F.” The EMBO Journal, vol. 17, no. 19, 1998, pp. 5811–5821., doi:10.1093/emboj/17.19.5811. </ref> |
== Biological Relevancy == | == Biological Relevancy == | ||
==='''Poly(A) Binding Protein's Evolution in plants'''=== | ==='''Poly(A) Binding Protein's Evolution in plants'''=== | ||
Revision as of 22:53, 29 March 2018
Poly(A) binding protein
Structure
| |||||||||||