User:Mark Macbeth/Sandbox2
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
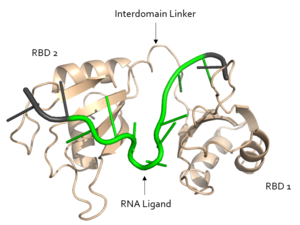
[[Image:Sex Lethal Protein Structural Overview with Labels.png|300px|left|thumb| '''Figure 1.''' Structural overview of Sxl. RNA ligand colored in green is recognized and bound, while RNA ligand colored in grey is not bound. Image created in PyMol. Structure shown is [https://www.rcsb.org/structure/1b7f PDB:1b7f].]] | [[Image:Sex Lethal Protein Structural Overview with Labels.png|300px|left|thumb| '''Figure 1.''' Structural overview of Sxl. RNA ligand colored in green is recognized and bound, while RNA ligand colored in grey is not bound. Image created in PyMol. Structure shown is [https://www.rcsb.org/structure/1b7f PDB:1b7f].]] | ||
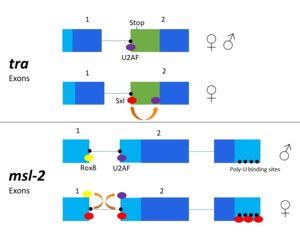
| - | The Sxl RNA splicing targets encode for the transformer (''tra'') and the male-sex lethal (''msl-2'') proteins. ''Tra'' is a splicing activator for the female developmental pathway, and ''msl-2'' modulates [http://en.wikipedia.org/wiki/X_chromosome X chromosome] application in male fruit flies. The mechanism for how Sxl targets these pathways differs slightly. In both mechanisms, Sxl occupies the 3' splice site and prevents [http://en.wikipedia.org/wiki/U2AF2 U2AF] from binding. This causes the U2AF splicing factor to bind at a downstream splice site encoding proteins in the female developmental pathway. In ''msl-2'' targeting, Sxl also blocks the binding of another regulatory splicing factor, Rox8, and the [http://en.wikipedia.org/wiki/SnRNP U1 snRNP] at the 5’ splice site<ref name="Penalva">. Sxl can also control its own splicing pattern to conserve female expression. It does so by binding to [http://en.wikipedia.org/wiki/Exon Exon] 3 of its own RNA and creating an RNP complex to eliminate this exon. After removal of Exon 3, Sxl becomes active and female expression is maintained. | + | The Sxl RNA splicing targets encode for the transformer (''tra'') and the male-sex lethal (''msl-2'') proteins. ''Tra'' is a splicing activator for the female developmental pathway, and ''msl-2'' modulates [http://en.wikipedia.org/wiki/X_chromosome X chromosome] application in male fruit flies. The mechanism for how Sxl targets these pathways differs slightly. In both mechanisms, Sxl occupies the 3' splice site and prevents [http://en.wikipedia.org/wiki/U2AF2 U2AF] from binding. This causes the U2AF splicing factor to bind at a downstream splice site encoding proteins in the female developmental pathway. In ''msl-2'' targeting, Sxl also blocks the binding of another regulatory splicing factor, Rox8, and the [http://en.wikipedia.org/wiki/SnRNP U1 snRNP] at the 5’ splice site<ref name="Penalva"/>. Sxl can also control its own splicing pattern to conserve female expression. It does so by binding to [http://en.wikipedia.org/wiki/Exon Exon] 3 of its own RNA and creating an RNP complex to eliminate this exon. After removal of Exon 3, Sxl becomes active and female expression is maintained. |
== Structure == | == Structure == | ||
Revision as of 18:03, 30 March 2018
| |||||||||||
Additional Reading
For more information on the U2AF splicing factor.
Relevance
As Sxl functions as a splicing repressor, it may give insight into the effects of varying mechanisms of alternate splicing both in flies and other species. Sxl may also lead to understanding of human alternative splicing factors. As an RNA binding protein, research regarding Sxl may contribute to the understanding of enzymes with RNA recognition motifs. The Sxl RNP motif of RBD1 is also conserved in the ELAV family of proteins[1].
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999 Apr 15;398(6728):579-85. PMID:10217141 doi:10.1038/19242
- ↑ 2.0 2.1 Penalva LO, Sanchez L. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev. 2003 Sep;67(3):343-59, table of contents. PMID:12966139
- ↑ 3.0 3.1 3.2 3.3 3.4 Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336. doi: 10.1146/annurev.biochem.72.121801.161720., Epub 2003 Feb 27. PMID:12626338 doi:http://dx.doi.org/10.1146/annurev.biochem.72.121801.161720