We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Jmol/Visualizing membrane position
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
Viewing the model in [[FirstGlance in Jmol]], you can use ''Hydrophobic/Polar'' (Views tab) to visualize the domain with a hydrophobic surface. Touching or clicking on atoms at the domain boundaries will identify those amino acids. If you wish, you can center a particular residue, then zoom in, and see atomic detail with ''Vines/Sticks'' (Views tab). | Viewing the model in [[FirstGlance in Jmol]], you can use ''Hydrophobic/Polar'' (Views tab) to visualize the domain with a hydrophobic surface. Touching or clicking on atoms at the domain boundaries will identify those amino acids. If you wish, you can center a particular residue, then zoom in, and see atomic detail with ''Vines/Sticks'' (Views tab). | ||
| + | |||
| + | For 5LiL, I chose thr607.og1 and leu250.cd1. "OG1" means "oxygen, gamma one", namely a sidechain oxygen that is 2 bonds away from the alpha carbon. CD1: carbon, delta one. It would have worked equally well to use the entire residues, simply thr607 and leu250. | ||
| + | |||
| + | ===Generate the Cylinder=== | ||
| + | |||
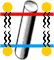
| + | To generate the cylinder in the [[SAT]], click on the button below JSmol ''Advanced: Open JSmol Console''. The command to generate the cylinder will have this form: | ||
| + | <pre>draw cyl1 cylinder diameter 100.0 color gray (thr607) (leu250)</pre> | ||
| + | |||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 19:30, 30 March 2018
| |||||||||||
References
- ↑ 1.0 1.1 Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012 Jan;40(Database issue):D370-6. doi: 10.1093/nar/gkr703., Epub 2011 Sep 2. PMID:21890895 doi:http://dx.doi.org/10.1093/nar/gkr703