User:Neel Bhagat/Sandbox 1
From Proteopedia
| Line 9: | Line 9: | ||
<StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | <StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | ||
=== Structure Determination === | === Structure Determination === | ||
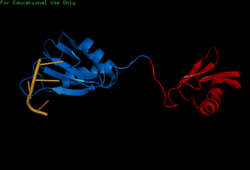
| - | [[Image:BothChains.png| | + | [[Image:BothChains.png|250px|right|thumb|'''Figure 1.''' This is a figure legend. Neel was here.]] |
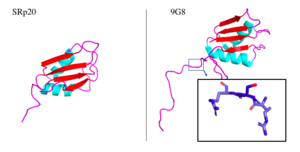
Attempts to determine the structure of native SRP20 were largely unsuccessful due to the low solubility of the protein. This is likely due to the hydrophobic core of the RRM and exposed hydrophobic residues for RNA recognition on the β-sheets. As a solution, researchers removed the SR domain from the C terminus, leaving only the SRP20 RRM and a small arginine rich segment at the C terminus, then fused with a soluble <scene name='78/782597/Imager0/2'>IgG binding domain</scene> of Streptococcal protein G to the N terminus of the protein, providing the first published structure of the SRP20 RRM via NMR3. However, the solution of the structure via NMR, in addition to fusion with a globular tag, results in multiple possible conformations of the protein, meaning measurements such as bond angles, lengths, and substrate interactions are variable. Further, information concerning structural aspects of the SR domain are still limited to experimental data of protein function with certain mutations or deletions, and by comparison to sister proteins such as 9G8. To date, structure of the SR domain or the protein without the globular tag have not been solved, nor has a crystal structure for any part of the protein been determined. | Attempts to determine the structure of native SRP20 were largely unsuccessful due to the low solubility of the protein. This is likely due to the hydrophobic core of the RRM and exposed hydrophobic residues for RNA recognition on the β-sheets. As a solution, researchers removed the SR domain from the C terminus, leaving only the SRP20 RRM and a small arginine rich segment at the C terminus, then fused with a soluble <scene name='78/782597/Imager0/2'>IgG binding domain</scene> of Streptococcal protein G to the N terminus of the protein, providing the first published structure of the SRP20 RRM via NMR3. However, the solution of the structure via NMR, in addition to fusion with a globular tag, results in multiple possible conformations of the protein, meaning measurements such as bond angles, lengths, and substrate interactions are variable. Further, information concerning structural aspects of the SR domain are still limited to experimental data of protein function with certain mutations or deletions, and by comparison to sister proteins such as 9G8. To date, structure of the SR domain or the protein without the globular tag have not been solved, nor has a crystal structure for any part of the protein been determined. | ||
== Splicing Activity == | == Splicing Activity == | ||
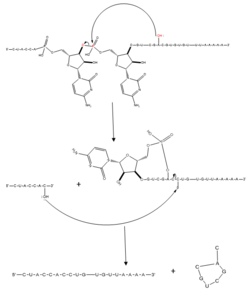
| - | [[Image:splicingmech.png| | + | [[Image:splicingmech.png|250px|left|thumb|'''Figure 2.''' Regular splicing mechanism for eukaryotes. Free 3’OH nucleophile of adenosine in intron attacks phosphorus of phosphate creating a ring structure intron known as a lariat. The free 2’OH in the 5’ splice site (red) acts as the nucleophile to attack the phosphate of the first nucleotide in the 3’ splice site (red) to release the lariat intron. The products include the final modified RNA sequence and the lariat which will be recycled.]] |
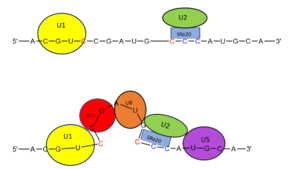
| - | The splicing mechanism for SRp20 follows the normal eukaryotic mechanism, in which five different small nuclear ribonucleoproteins (snRNPs) bring the splice sites together in order to start the reaction ( | + | [[Image:snurpmech.png|299px|right|thumb|'''Figure 3.''' SRp20 works with U2 snRNP: Five snRNPs are needed in the eukaryotic splicing mechanism to facilitate the reaction. The U2 snRNP must attach to the 3’ splice site to bring together the 5’ and 3’ splice sites (red). SRp20 facilitates binding of U2 to the 3’ splice site by binding to the exonic splicing enhancer sequence (blue) in the RNA at the backbone. U2 must bind before the other snRNPs can bind to continue the mechanism.]] |
| + | The splicing mechanism for SRp20 follows the normal eukaryotic mechanism, in which five different small nuclear ribonucleoproteins (snRNPs) bring the splice sites together in order to start the reaction (Figure 2). SRp20 and other SR proteins interact with the RNA ligand at the exonic splicing enhancer sequence at the beginning of the 3’ splice site adjacent to the intron being removed. SRp20 facilitates the interaction of the U2 snRNP with the RNA to continue the mechanism (Figure 3). | ||
Revision as of 16:43, 2 April 2018
Contents |
Introduction
Overview
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other eukaryotes. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM).
History
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. Alternative splicing allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins (hnRNPs). The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the SF2 (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). An identical protein, called X16, was discovered in an earlier paper studying different genes that change expression during B-cell development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013).
| |||||||||||