User:Neel Bhagat/Sandbox 1
From Proteopedia
| Line 41: | Line 41: | ||
=== Cancer === | === Cancer === | ||
| - | SRp20 has been linked to [https://en.wikipedia.org/wiki/Cancer cancer] in many instances, as well as other AS proteins. SRp20 has been seen to activate the AS of the [http://proteopedia.org/wiki/index.php/CD44 CD44 adhesion molecule.] SRp20 facilitates the splicing of exon v9, one important for function of CD44. Loss of function of SRp20 leading to loss of function of CD44 will lead to loss of “stickiness” of cells, a way that cancer cells can spread to other areas of the body. SRp20 can also affect the alternative splicing of [https://en.wikipedia.org/wiki/Oncogene oncogenes] and [https://en.wikipedia.org/wiki/Tumor_suppressor_gene tumor suppressors]. Expression of the signaling pathway for SRp20 translation is directly triggered by oncogenic signalling pathways. For example, the Wnt pathway that is associated with cancer development also drives further expression of SRp20 and other SR proteins (7). These proteins aid in cell growth and proliferation, in that they increase expression of proteins they are involved in splicing. One study found that silencing SRp20 slowed cell proliferation, further supporting the idea that SRp20 helps with this process (8). Cell cycle regulator proteins FoxM1, Cdc25B, and PLK1 are alternatively spliced by SRp20, specifically regulating the G2/M cycles. These proteins are also associated with regulation of cell apoptosis. When these genes are overexpressed due to overexpressed SRp20, cells will no longer be able to apoptose when they are damaged, and the cell cycle will continue without regulation, causing overactive cell proliferation and eventually cancer. SRp20 has been seen to be overexpressed in ovarian, lung, breast, stomach, skin, bladder, colon, liver, thyroid, and kidney cancer tissue (9). SRp20 also regulates genes associated with cellular senescence, or cellular immortality, which contributes to cancer formation. Specifically, SRp20 alternatively splices the TP53 gene, which generates the p53 senescence protein. | + | SRp20 has been linked to [https://en.wikipedia.org/wiki/Cancer cancer] in many instances, as well as other AS proteins. SRp20 has been seen to activate the AS of the [http://proteopedia.org/wiki/index.php/CD44 CD44 adhesion molecule.] SRp20 facilitates the splicing of exon v9, one important for function of CD44. Loss of function of SRp20 leading to loss of function of CD44 will lead to loss of “stickiness” of cells, a way that cancer cells can spread to other areas of the body. SRp20 can also affect the alternative splicing of [https://en.wikipedia.org/wiki/Oncogene oncogenes] and [https://en.wikipedia.org/wiki/Tumor_suppressor_gene tumor suppressors]. Expression of the signaling pathway for SRp20 translation is directly triggered by oncogenic signalling pathways. For example, the Wnt pathway that is associated with cancer development also drives further expression of SRp20 and other SR proteins (7). These proteins aid in cell growth and proliferation, in that they increase expression of proteins they are involved in splicing. One study found that silencing SRp20 slowed cell proliferation, further supporting the idea that SRp20 helps with this process (8). Cell cycle regulator proteins FoxM1, Cdc25B, and PLK1 are alternatively spliced by SRp20, specifically regulating the G2/M cycles. These proteins are also associated with regulation of cell apoptosis. When these genes are overexpressed due to overexpressed SRp20, cells will no longer be able to apoptose when they are damaged, and the cell cycle will continue without regulation, causing overactive cell proliferation and eventually cancer. SRp20 has been seen to be overexpressed in ovarian, lung, breast, stomach, skin, bladder, colon, liver, thyroid, and kidney cancer tissue (9). SRp20 also regulates genes associated with cellular senescence, or cellular immortality, which contributes to cancer formation. Specifically, SRp20 alternatively splices the [https://ghr.nlm.nih.gov/gene/TP53 TP53] gene, which generates the [http://proteopedia.org/wiki/index.php/P53 p53] senescence protein. |
| + | |||
=== Neurological Disorders === | === Neurological Disorders === | ||
| - | Along with cancer, SRp20 mutations have been linked to Alzheimers, a neurodegenerative disorder. SRp20 is involved in AS of a wide array of RNAs, including that of the TRKB gene to generate TrkB-Shc transcripts that are involved in generation of the disorder. SRp20 also promotes exclusion of exon 10 in the TAU gene, a gene important in establishing microtubules in axons or transport processes.Underexpression of SRp20 results in dysfunction of the TAU gene, less microtubule functionality, and the brain deterioration characteristic of Alzheimers (10, 7). | + | Along with cancer, SRp20 mutations have been linked to [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimers], a neurodegenerative disorder. SRp20 is involved in AS of a wide array of RNAs, including that of the [https://en.wikipedia.org/wiki/Tropomyosin_receptor_kinase_B TRKB] gene to generate TrkB-Shc transcripts that are involved in generation of the disorder. SRp20 also promotes exclusion of exon 10 in the TAU gene, a gene important in establishing microtubules in axons or transport processes.Underexpression of SRp20 results in dysfunction of the TAU gene, less microtubule functionality, and the brain deterioration characteristic of Alzheimers (10, 7). |
Revision as of 18:00, 2 April 2018
Contents |
Introduction
Overview
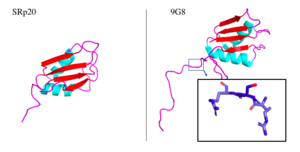
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other eukaryotes. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM).
History
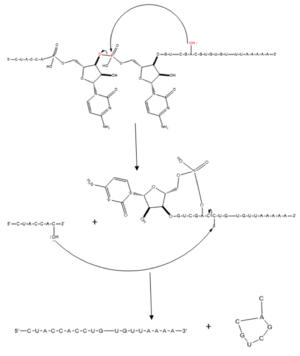
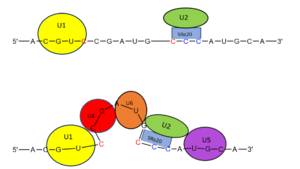
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. Alternative splicing allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins (hnRNPs). The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the SF2 (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). An identical protein, called X16, was discovered in an earlier paper studying different genes that change expression during B-cell development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013). The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006).
| |||||||||||