User:David Ryskamp/Sandbox1
From Proteopedia
| Line 35: | Line 35: | ||
===Recognition of the poly(A) tail=== | ===Recognition of the poly(A) tail=== | ||
| - | Polyadenylation of an mRNA involves the recognition of the 5’-AAUAAA-3’ consensus site, the cleavage downstream of the consensus site, and then the addition of adenines by Poly(A) Polymerase to the 3’ end. The newly added poly(A) tail is | + | Polyadenylation of an mRNA involves the recognition of the 5’-AAUAAA-3’ consensus site, the cleavage downstream of the consensus site, and then the addition of adenines by Poly(A) Polymerase to the 3’ end. The newly added poly(A) tail is associated with the PABP. PABP requires 11-12 adenosines in order to bind. PABP and the bound Poly(A) tail work together to stabilize mRNA by preventing exo-ribonucleolytic degradation,¹ thereby guiding the mRNA molecule into the translation pathway. Upon mRNA poly(A) recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor eIF4G. Mutations of Arg→Ala and Lys→Ala in human eIF4G and in yeast extracts decrease the rate of translation initiation and destabilizing the interactions with PABP, indicating that basic residues are essential to the interaction with PABP.¹ |
| + | |||
<table><tr><td colspan='2'> | <table><tr><td colspan='2'> | ||
| Line 49: | Line 50: | ||
===mRNA Stabilization=== | ===mRNA Stabilization=== | ||
| - | PABP prevents the deadenylation and decapping of the mRNA, serving as a source of stabilization. Poly(A) ribonuclease (PARN) work to deadenylate mRNA, but the presence of PABP prevents its activity | + | PABP prevents the deadenylation and decapping of the mRNA, serving as a source of stabilization. Poly(A) ribonuclease (PARN) work to deadenylate mRNA, but the presence of PABP prevents its activity. The PABP protein is able to protect mRNA degradation through the complex that it forms with the elongation initiation factors, which prevent deadenylation and decapping due to their presence.¹ This has been verified by the presence of deadenylation products and the comparative size of PABP footprints. There is some evidence indicating that PABP is involved in the prevention of endonucleolytic cleavage; however, only a small amount of mRNA is degraded from endonucleolytic cleavage, so it is not widely researched.¹ |
===Eukaryotic Translation Initiation=== | ===Eukaryotic Translation Initiation=== | ||
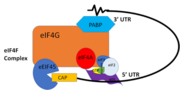
| - | + | PABP and mRNA complex aids in translation initiation under two proposed mechanisms. Within the two mechanisms, studies have highlighted the presence The “Closed Loop” Model entails the recognition of the 5’ 7-methyl-Guanosine cap by eIF4F, which is a ternary complex made up of a cap-binding protein (eIF4E) and RNA helicase (eIF4A) connected by the bridging protein (eIF4G).¹ Translation initiation is stimulated by the PABP bound to the poly(A) tail and its association with eIF4G.¹ The 5’ UTR is unwound by the elF4F complex, and ribosomes are recruited to create the initiation complex. The eIF4G protein then guides the 40S subunit to the start codon (AUG), which is followed by the binding 60S ribosomal subunit, creating the 80S initiation complex.¹ The association of the PABP and eIF4G gave rise to the name “closed loop.”¹ | |
| - | + | ||
| - | PABP and mRNA complex aids in translation initiation under two proposed mechanisms. Within the two mechanisms, studies have highlighted the presence The “Closed Loop” Model entails the recognition of the 5’ 7-methyl-Guanosine | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | The 5’ UTR is unwound by the elF4F complex, and ribosomes are recruited to create the initiation complex. The eIF4G protein then guides the 40S subunit to the start codon (AUG), which is followed by the binding 60S ribosomal subunit, creating the 80S initiation complex.¹ The association of the | + | |
| + | [[Image:closedlooper.png|thumb|350px "Figure 1:" Closed loop model of the eIF4F complex and PABP creating a loop out of the mRNA ]] | ||
| + | In more complex eukaryotic organisms, PABP indirectly stimulates translation via PAIP-1 (PABP interacting protein). A higher presence of PAIP-1 increases the rate of translation initiation, indicating another way to “close the loop.”¹ | ||
== Disease and Medical Relevance == | == Disease and Medical Relevance == | ||
Revision as of 21:32, 2 April 2018
Human Poly(A) Binding Protein (1CVJ)
| |||||||||||
References
1. Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print.
2. Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.
3. “Oculopharyngeal Muscular Dystrophy.” NORD (National Organization for Rare Disorders), rarediseases.org/rare-diseases/oculopharyngeal-muscular-dystrophy/.
4. Richard, Pascale, et al. “Correlation between PABPN1 Genotype and Disease Severity in Oculopharyngeal Muscular Dystrophy.” Neurology, vol. 88, no. 4, 2016, pp. 359–365., doi:10.1212/wnl.0000000000003554.
5. Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644