User:Mark Hoelzer/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
<StructureSection load='1a3n' size='400' side='right' caption='Hemoglobin based on 1a3n.pdb' scene=''> | <StructureSection load='1a3n' size='400' side='right' caption='Hemoglobin based on 1a3n.pdb' scene=''> | ||
| - | |||
| - | This is a default text for your page '''Mark Hoelzer/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | |||
| - | == Function == | ||
| - | |||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | |||
| - | </StructureSection> | ||
| - | == References == | ||
| - | <references/> | ||
Revision as of 21:35, 2 April 2018
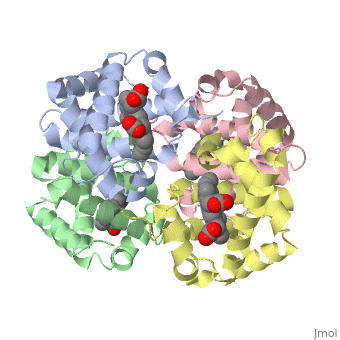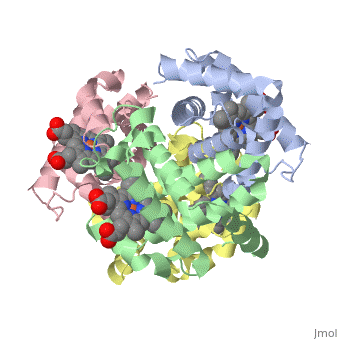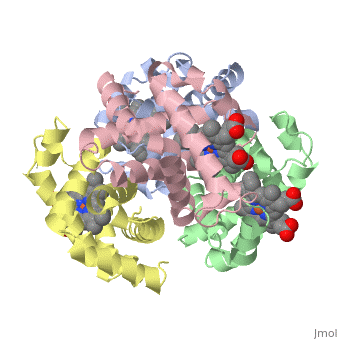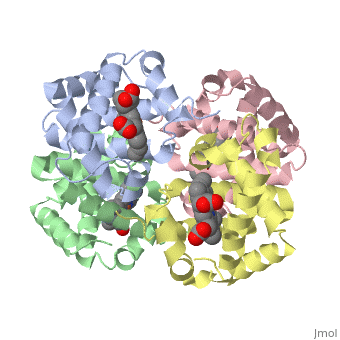
3D Printed Physical Model of Hemoglobin
The MSOE Center for BioMolecular Modeling uses 3D printing technology to create physical models of protein and molecular structures. Models function as thinking tools, making the invisible molecular world more tangible and comprehensible.
Shown to the right is a physical model of Hemoglobin, based on the structure 1a3n.pdb.
|