This is a default text for your page Ben Dawson/Sandbox1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Background
The Human Poly(A) Binding Protein (PABP) was discovered in 1973 by the use of a sedimentation profile detailing the RNase digestion differentiated the PABP protein.6 Attempts to purify the 75 kDa protein then followed. In 1983, then considered “poly(A)-organizing protein,” was determined and purified by molecular weight, ligand-binding affinity, and amounts found in cytoplasmic portions of cell with ability to bind to free poly(A).8 The protein’s structure was initially determined at 2.6 Å as mRNA and RRM 1 & 2 yielded high cocrystal quality with 1:1 protein and ligand.1 The protein has a homopolymeric structure, containing four RNA recognition motifs (RRMs), which are conserved.7 RRM1 and RRM2 are N-terminal domains that are connected by a linker.1 Opposed to their counterparts, RRM3 and RRM4 bind Poly-A RNA less tightly than RRM1 and RRM2.1
PABP is a mRNA binding protein that binds to the 3’ Poly(A) tail on mRNA. Through extensive Adenosine recognition by the RRMs of PABP, the protein is involved in three main functions: recognition of the 3’ Poly(A) tail, mRNA stabilization, and eukaryotic translation initiation. The contributions of controlling gene expression via different families of PABPs is not yet fully understood. PABP families are divided into nuclear and cytoplasmic.5 PABP1, which is predominantly cytoplasmic, is often referred to as PABP because it is the only form of PABP that has been extensively studied in its role with mRNA translation and stability.5
Structure
The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). It is comprised of four RNA recognition motifs (RRMs), which are highly conserved RNA-binding domains.¹ The RRM in PABP is found in over two hundred families of proteins across species, indicating that it is ancient.¹ RRM1 and 2 are examined in this article.
Each RRM has a four-stranded antiparallel beta sheet backed by two corresponding alpha helices.¹ mRNA poly-adenosine recognition is due to the presence of the conserved residues within the beta-sheet surface², which forms a -like pocket for the mRNA to bind. The beta-sheet flooring present in PABP interacts with the 3’ mRNA tail via a combination of van der Waals, aromatic stacking, and Hydrogen bonding. Through these interactions, PABP binds to 3’ poly-adenosine tail with a KD of 2-7 nM.5
Further, the RRM1/2 complex interacts with the mRNA's sugar-phosphate backbone, where 4 of the 8 mRNA adenosines interact electrostatically¹. Upon closer examination of the PABP structure, the protein contains loop-like domains that form the walls of the beta-sheet trough. Although these loop walls are present, no interaction occurs between the mRNA and these regions. We propose that these loops only keep unwanted cellular elements out of the binding pocket via hydrophobic and hydrophilic interactions, maintaining the protein's selectivity for mRNA. The structural elements highlighted consist of the RRM1/2 subunits, the linker domain, and the Poly(A) mRNA binding trough.
RNA Recognition Motifs (RRMs)
The are categorized into two RRMs: the n-terminus RRM1 (red) and c-terminus RRM2 (blue) are shown accordingly. The two RRMs are linked via an alpha-helix linker (green) that maintains the RRM1/2 complex that is the biological assembly and active form of PABP.
Poly-A RNA Binding Trough
Interactions
Adenosine Recognition via Hydrogen Bonding
|
|
| Adenosine Number | PABP residue | Atom on Protein |
| A2 | | side chain amine |
| A3 | | amine |
| A4 | | side chain amine and carbonyl |
| A4 | | side chain and N-term amine |
| A5 | | pyrimidine π-amine |
| A6 | | N-term amine and C-term OH |
| A6 | | N-term amine |
| A7 | | side chain OH |
| A7 | | side chain OH |
| A7 | | C-term carbonyl |
| A8 | | side chain OH |
| A9 | | side chain τ-amine |
Aromatic Stacking
Function
In eukaryotic mRNA translation, PABP recognizes the 3' Poly(A) tail via trough interactions determined above. While associated with the Poly(A) region, the complex then works together to stabilize the mRNA by preventing exoribonucleolytic degradation,¹ thereby guiding the mRNA molecule into the translation pathway via interactions with translation initiation factor gG.
Recognition of the poly(A) tail
Polyadenylation of an mRNA involves the recognition of the 5’-AAUAAA-3’ consensus site, the cleavage downstream of the consensus site, and then the addition of adenines by Poly(A) Polymerase to the 3’ end. The newly added poly(A) tail is associated with the PABP. PABP requires 11-12 adenosines in order to bind. PABP and the bound Poly(A) tail work together to stabilize mRNA by preventing exo-ribonucleolytic degradation,¹ thereby guiding the mRNA molecule into the translation pathway. Upon mRNA poly(A) recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor eIF4G. Mutations of Arg→Ala and Lys→Ala in human eIF4G and in yeast extracts decrease the rate of translation initiation and destabilizing the interactions with PABP, indicating that basic residues are essential to the interaction with PABP.¹
mRNA Stabilization
PABP prevents the deadenylation and decapping of the mRNA, serving as a source of stabilization. Poly(A) ribonuclease (PARN) work to deadenylate mRNA, but the presence of PABP prevents its activity. The PABP protein is able to protect mRNA degradation through the complex that it forms with the elongation initiation factors, which prevent deadenylation and decapping due to their presence.¹ This has been verified by the presence of deadenylation products and the comparative size of PABP footprints. There is some evidence indicating that PABP is involved in the prevention of endonucleolytic cleavage; however, only a small amount of mRNA is degraded from endonucleolytic cleavage, so it is not widely researched.¹
Eukaryotic Translation Initiation
PABP and mRNA complex aids in translation initiation under two proposed mechanisms. Within the two mechanisms, studies have highlighted the presence The “Closed Loop” Model entails the recognition of the 5’ 7-methyl-Guanosine cap by [ https://en.wikipedia.org/wiki/Eukaryotic_initiation_factor_4F eIF4F], which is a ternary complex made up of a cap-binding protein (eIF4E) and RNA helicase (eIF4A) connected by the bridging protein (eIF4G).¹ Translation initiation is stimulated by the PABP bound to the poly(A) tail and its association with eIF4G.¹ The 5’ UTR is unwound by the elF4F complex, and ribosomes are recruited to create the initiation complex. The eIF4G protein then guides the 40S subunit to the start codon (AUG), which is followed by the binding 60S ribosomal subunit, creating the 80S initiation complex.¹ The association of the PABP and eIF4G gave rise to the name “closed loop.”¹
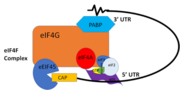
350px "Figure 1:" Closed loop model of the eIF4F complex and PABP creating a loop out of the mRNA
In more complex eukaryotic organisms, PABP indirectly stimulates translation via PAIP-1 (PABP interacting protein). A higher presence of PAIP-1 increases the rate of translation initiation, indicating another way to “close the loop.”¹
Disease and Medical Relevance
===[https://rarediseases.info.nih.gov/diseases/7245/oculopharyngeal-muscular-dystrophy
https://ghr.nlm.nih.gov/gene/PABPN1#resources Oculopharyngeal muscular dystrophy (OPMD)]===
Oculopharyngeal muscular dystrophy, or OPMD, is an autosomal dominant late-onset disease.³ It’s characterized by the myopathy of the eyelids and the throat. The symptoms entail eye-drooping and difficulty swallowing. There are two types of OPMD: autosomal dominant and recessive, both originating from the mutation of the PABP nuclear 1 (PABPN1) gene located on the long arm of chromosome 14.³ This mutation results in an abnormally long polyalanine tract, 11-18 alanines, opposed to the normal 10.³ Patients with longer PABPN1 expansion (more alanines) are on average diagnosed at an earlier in life than patients with a shorter expansion; therefore, expansion size plays a role in OPMD severity and progression. 4
The mutation results in PABPN1 forming clumps in muscle cells that can’t be degraded.³ It’s suspected that this is a source of cell death for effected cells, however, it has not been concluded why this mutation only affects certain muscle cells.
Studies on Mutations
Studies conducted on Drosophila are common due to 75% conservation between human and Drosophila genomes. Drosophila only encode one cytoplasmic PABP, and its deletion results in embryonic lethality.5 Similarly, in Caenorhabditis elegans, which have two cytoplasmic PABPs, display 50-80% embryonic lethality with the introduction of an RNAi to one of these PABPs.5
Relevance
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.

