User:Ben Dawson/Sandbox1
From Proteopedia
| Line 12: | Line 12: | ||
== Structure == | == Structure == | ||
| - | The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). It is comprised of four RNA recognition motifs (RRMs), which are highly conserved RNA-binding domains.¹ The RRM in PABP is found in over two hundred families of proteins across species, indicating that it is ancient.¹ RRM1 and 2 are examined in this article. | + | The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). It is comprised of four [https://en.wikipedia.org/wiki/RNA_recognition_motif&sa=D&ust=1522708537701000&usg=AFQjCNGga6XMrUjL5zum75ip4i4Jzlwa0g RNA recognition motifs] (RRMs), which are highly conserved RNA-binding domains.¹ The RRM in PABP is found in over two hundred families of proteins across species, indicating that it is ancient.¹ RRM1 and 2 are examined in this article. |
Each RRM has a four-stranded antiparallel beta sheet backed by two corresponding alpha helices.¹ mRNA poly-adenosine recognition is due to the presence of the conserved residues within the beta-sheet surface², which forms a <scene name='78/782616/Trough2/1'>trough</scene>-like pocket for the mRNA to bind. The beta-sheet flooring present in PABP interacts with the 3’ mRNA tail via a combination of van der Waals, aromatic stacking, and Hydrogen bonding. Through these interactions, PABP binds to 3’ poly-adenosine tail with a KD of 2-7 nM.5 | Each RRM has a four-stranded antiparallel beta sheet backed by two corresponding alpha helices.¹ mRNA poly-adenosine recognition is due to the presence of the conserved residues within the beta-sheet surface², which forms a <scene name='78/782616/Trough2/1'>trough</scene>-like pocket for the mRNA to bind. The beta-sheet flooring present in PABP interacts with the 3’ mRNA tail via a combination of van der Waals, aromatic stacking, and Hydrogen bonding. Through these interactions, PABP binds to 3’ poly-adenosine tail with a KD of 2-7 nM.5 | ||
| Line 38: | Line 38: | ||
===mRNA Stabilization=== | ===mRNA Stabilization=== | ||
| - | PABP prevents the deadenylation and decapping of the mRNA, serving as a source of stabilization. Poly(A) ribonuclease (PARN) work to deadenylate mRNA, but the presence of PABP prevents its activity; this has been verified by the presence of deadenylation products in the size of PABP footprints. Further, the PABP protein is able to protect mRNA degradation through the complex that it forms with the elongation initiation factors, which prevent deadenylation and decapping due to their presence.¹ There is some evidence indicating that PABP is involved in the prevention of endonucleolytic cleavage; however, only a small amount of mRNA is degraded from endonucleolytic cleavage, so it is not widely researched.¹ | + | PABP prevents the deadenylation and decapping of the mRNA, serving as a source of stabilization. Poly(A) ribonuclease ([https://en.wikipedia.org/wiki/Poly(A)specific_ribonuclease&sa=D&ust=1522708205779000&usg=AFQjCNEyJO40-Jktj58e0CQnGJnaPjll3w PARN]) work to deadenylate mRNA, but the presence of PABP prevents its activity; this has been verified by the presence of deadenylation products in the size of PABP footprints. Further, the PABP protein is able to protect mRNA degradation through the complex that it forms with the elongation initiation factors, which prevent deadenylation and decapping due to their presence.¹ There is some evidence indicating that PABP is involved in the prevention of endonucleolytic cleavage; however, only a small amount of mRNA is degraded from endonucleolytic cleavage, so it is not widely researched.¹ |
===Eukaryotic Translation Initiation=== | ===Eukaryotic Translation Initiation=== | ||
Upon mRNA Poly(A) recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor eIF4G. Protein eIF4G actually interacts with PABP's dorsal side (under the trough) <scene name='78/782616/Residues_involved_translations/4'>hydrophobic and acidic residues</scene> that stimulate the interaction between the two proteins. These specific residues are phylogenetically conserved among all PABPs, and therefore significant in the protein's function and interaction with eIF4G. | Upon mRNA Poly(A) recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor eIF4G. Protein eIF4G actually interacts with PABP's dorsal side (under the trough) <scene name='78/782616/Residues_involved_translations/4'>hydrophobic and acidic residues</scene> that stimulate the interaction between the two proteins. These specific residues are phylogenetically conserved among all PABPs, and therefore significant in the protein's function and interaction with eIF4G. | ||
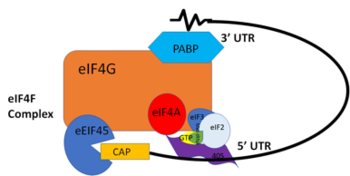
| - | PABP and mRNA complex aids in translation initiation under two proposed mechanisms. Within the two mechanisms, studies have highlighted the presence The “Closed Loop” Model entails the recognition of the 5’ 7-methyl-Guanosine Cap by eIF4F, which is a ternary complex made up of a cap-binding protein (eIF4E) and RNA helicase (eIF4A) connected by the bridging protein (eIF4G).¹ Translation initiation is stimulated by the PABP bound to the Poly-A tail and its association with eIF4G.¹ With respect to this stimulation, mutations of Arg→Ala and Lys→Ala in human eIF4G decrease the rate of translation initiation and destabilizing the interactions with PABP, indicating that basic residues are essential to the interaction with PABP.¹ | + | PABP and mRNA complex aids in translation initiation under two proposed mechanisms. Within the two mechanisms, studies have highlighted the presence The “Closed Loop” Model entails the recognition of the 5’ 7-methyl-Guanosine Cap by [https://en.wikipedia.org/wiki/Eukaryotic_initiation_factor_4F&sa=D&ust=1522708342158000&usg=AFQjCNHP-IfV_6Q-kPtQNHb96jSKIagURg eIF4F], which is a ternary complex made up of a cap-binding protein ([https://en.wikipedia.org/wiki/EIF4E&sa=D&ust=1522708379234000&usg=AFQjCNHIxw0niE3JON1U6HIJho70lFh7kA eIF4E]) and RNA helicase ([https://en.wikipedia.org/wiki/EIF4A&sa=D&ust=1522708420475000&usg=AFQjCNHqeHGnKw1Jdatgb-zU3CXCVl54zQ eIF4A]) connected by the bridging protein (eIF4G).¹ Translation initiation is stimulated by the PABP bound to the Poly-A tail and its association with eIF4G.¹ With respect to this stimulation, mutations of Arg→Ala and Lys→Ala in human eIF4G decrease the rate of translation initiation and destabilizing the interactions with PABP, indicating that basic residues are essential to the interaction with PABP.¹ |
[[Image:closedlooper.png|thumb|350px]] | [[Image:closedlooper.png|thumb|350px]] | ||
| - | The 5’ UTR is unwound by the elF4F complex, and ribosomes are recruited to create the initiation complex. The eIF4G protein then guides the 40S subunit to the start codon (AUG), which is followed by the binding 60S ribosomal subunit, creating the 80S initiation complex.¹ The association of the PolyA binding protein and eIF4G gave rise to the name “closed loop.”¹ In more complex eukaryotic organisms, PABP indirectly stimulates translation via PAIP-1 (PABP interacting protein). A higher presence of PAIP-1 increases the rate of translation initiation, indicating another way to “close the loop.”¹ | + | The 5’ UTR is unwound by the elF4F complex, and ribosomes are recruited to create the initiation complex. The eIF4G protein then guides the 40S subunit to the start codon (AUG), which is followed by the binding 60S ribosomal subunit, creating the 80S initiation complex.¹ The association of the PolyA binding protein and eIF4G gave rise to the name “closed loop.”¹ In more complex eukaryotic organisms, PABP indirectly stimulates translation via [https://en.wikipedia.org/wiki/PAIP1&sa=D&ust=1522708477059000&usg=AFQjCNHuoon5t1sVB4yJSQQxnzb_fJp0Xg PAIP-1] (PABP interacting protein). A higher presence of PAIP-1 increases the rate of translation initiation, indicating another way to “close the loop.”¹ |
| Line 55: | Line 55: | ||
===Oculopharyngeal muscular dystrophy (OPMD)=== | ===Oculopharyngeal muscular dystrophy (OPMD)=== | ||
| - | Oculopharyngeal muscular dystrophy, or OPMD, is an autosomal dominant late-onset disease.³ It’s characterized by the myopathy of the eyelids and the throat. The symptoms entail eye-drooping and difficulty swallowing. There are two types of OPMD: autosomal dominant and recessive, both originating from the mutation of the PABP nuclear 1 (PABPN1) gene located on the long arm of chromosome 14.³ This mutation results in an abnormally long polyalanine tract, 11-18 alanines, opposed to the normal 10.³ Patients with longer PABPN1 expansion (more alanines) are on average diagnosed at an earlier in life than patients with a shorter expansion; therefore, expansion size plays a role in OPMD severity and progression. 4 | + | Oculopharyngeal muscular dystrophy, or [https://rarediseases.info.nih.gov/diseases/7245/oculopharyngeal-muscular-dystrophy&sa=D&ust=1522706780528000&usg=AFQjCNE2EjxSdJXWA4SQA-1Da_EIugGlFA OPMD], is an autosomal dominant late-onset disease.³ It’s characterized by the myopathy of the eyelids and the throat. The symptoms entail eye-drooping and difficulty swallowing. There are two types of OPMD: autosomal dominant and recessive, both originating from the mutation of the PABP nuclear 1 ([https://en.wikipedia.org/wiki/PABPN1&sa=D&ust=1522708334189000&usg=AFQjCNHyfwTlpNLQZwHpmV8vlhMi9CXx_g PABPN1]) gene located on the long arm of chromosome 14.³ This mutation results in an abnormally long polyalanine tract, 11-18 alanines, opposed to the normal 10.³ Patients with longer PABPN1 expansion (more alanines) are on average diagnosed at an earlier in life than patients with a shorter expansion; therefore, expansion size plays a role in OPMD severity and progression. 4 |
The mutation results in PABPN1 forming clumps in muscle cells that can’t be degraded.³ It’s suspected that this is a source of cell death for effected cells, however, it has not been concluded why this mutation only affects certain muscle cells. | The mutation results in PABPN1 forming clumps in muscle cells that can’t be degraded.³ It’s suspected that this is a source of cell death for effected cells, however, it has not been concluded why this mutation only affects certain muscle cells. | ||
===Studies on Mutations=== | ===Studies on Mutations=== | ||
| - | Studies conducted on Drosophila are common due to 75% conservation between human and Drosophila genomes. Drosophila only encode one cytoplasmic PABP, and its deletion results in embryonic lethality.5 Similarly, in Caenorhabditis elegans, which have two cytoplasmic PABPs, display 50-80% embryonic lethality with the introduction of an RNAi to one of these PABPs.5 | + | Studies conducted on [https://en.wikipedia.org/wiki/Drosophila&sa=D&ust=1522708285193000&usg=AFQjCNHomZRiWyjtUMoBsBTrmQSRq3qUCg Drosophila] are common due to 75% conservation between human and Drosophila genomes. Drosophila only encode one cytoplasmic PABP, and its deletion results in embryonic lethality.5 Similarly, in [https://en.wikipedia.org/wiki/Caenorhabditis_elegans&sa=D&ust=1522708302132000&usg=AFQjCNE7XKhmyTGndBkUyYZwxRlCBzju4g Caenorhabditis elegans], which have two cytoplasmic PABPs, display 50-80% embryonic lethality with the introduction of an RNAi to one of these PABPs.5 |
| Line 81: | Line 81: | ||
5. Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125. | 5. Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125. | ||
| + | |||
| + | |||
<references/> | <references/> | ||
Revision as of 21:54, 2 April 2018
Human Poly(A) Binding Protein (1CVJ)
| |||||||||||
References
1. Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print.
2. Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.
3. “Oculopharyngeal Muscular Dystrophy.” NORD (National Organization for Rare Disorders), rarediseases.org/rare-diseases/oculopharyngeal-muscular-dystrophy/.
4. Richard, Pascale, et al. “Correlation between PABPN1 Genotype and Disease Severity in Oculopharyngeal Muscular Dystrophy.” Neurology, vol. 88, no. 4, 2016, pp. 359–365., doi:10.1212/wnl.0000000000003554.
5. Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644