User:Neel Bhagat/Sandbox 1
From Proteopedia
| Line 53: | Line 53: | ||
== References == | == References == | ||
1. [https://ac.els-cdn.com/S0168952501026269/1-s2.0-S0168952501026269-main.pdf?_tid=26ec5bda-f99a-4419-a621-823fa5d8a946&acdnat=1522368120_117c3ca278bff25bd0d673f8e4a3c185 Cáceres JF and Kornblihtt AR. 2002. “Alternative splicing: multiple control mechanisms and involvement in human disease.” TRENDS in Genetics; 18(4):186-193.] | 1. [https://ac.els-cdn.com/S0168952501026269/1-s2.0-S0168952501026269-main.pdf?_tid=26ec5bda-f99a-4419-a621-823fa5d8a946&acdnat=1522368120_117c3ca278bff25bd0d673f8e4a3c185 Cáceres JF and Kornblihtt AR. 2002. “Alternative splicing: multiple control mechanisms and involvement in human disease.” TRENDS in Genetics; 18(4):186-193.] | ||
| + | |||
2. [http://mcb.asm.org/content/18/9/4977.full.pdf+html Lou H, Neugebauer KM, Gagel RF, and Berget SM. 1998. “Regulation of Alternative Polyadenylation by U1 snRNPs and SRp20.” Molecular and Cellular Biology; 18(9):4977-4985.] | 2. [http://mcb.asm.org/content/18/9/4977.full.pdf+html Lou H, Neugebauer KM, Gagel RF, and Berget SM. 1998. “Regulation of Alternative Polyadenylation by U1 snRNPs and SRp20.” Molecular and Cellular Biology; 18(9):4977-4985.] | ||
| + | |||
3. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1630407/pdf/7601385a.pdf Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golocanov AP, et. al. 2006. “Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.” The EMBO Journal; 25(21):5126-5137.] | 3. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1630407/pdf/7601385a.pdf Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golocanov AP, et. al. 2006. “Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.” The EMBO Journal; 25(21):5126-5137.] | ||
| + | |||
4. [http://genesdev.cshlp.org/content/6/5/837.short Zahler, A. M., Lane, W. S., Stolk, J. A., & Roth, M. B. 1992. “SR proteins: a conserved family of pre-mRNA splicing factors.” Genes & development; 6(5):837-847.] | 4. [http://genesdev.cshlp.org/content/6/5/837.short Zahler, A. M., Lane, W. S., Stolk, J. A., & Roth, M. B. 1992. “SR proteins: a conserved family of pre-mRNA splicing factors.” Genes & development; 6(5):837-847.] | ||
| + | |||
5. [https://www.sciencedirect.com/science/article/pii/S0006291X13008097 Corbo C, Orrù S, and Salvatore F. 2013. “SRp20: An overview of its role in human diseases.” Biochemical and Biophysical Research Communications; 436(1):1-5.] | 5. [https://www.sciencedirect.com/science/article/pii/S0006291X13008097 Corbo C, Orrù S, and Salvatore F. 2013. “SRp20: An overview of its role in human diseases.” Biochemical and Biophysical Research Communications; 436(1):1-5.] | ||
| + | |||
6. [https://onlinelibrary.wiley.com/doi/epdf/10.1002/pmic.201100370 Corbo C, Orrù S, Gemei M, Di Noto R, Mirabelli P, et. al. “Protein cross-talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumerigenicity.” Proteomics; 12(12):2045-2059.] | 6. [https://onlinelibrary.wiley.com/doi/epdf/10.1002/pmic.201100370 Corbo C, Orrù S, Gemei M, Di Noto R, Mirabelli P, et. al. “Protein cross-talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumerigenicity.” Proteomics; 12(12):2045-2059.] | ||
| + | |||
7. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3005347/ Jia R, Li C, McCoy JP, Deng C, and Zheng Z. 2010. “SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance.” International Journal of Biological Sciences; 6(7):806-826.] | 7. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3005347/ Jia R, Li C, McCoy JP, Deng C, and Zheng Z. 2010. “SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance.” International Journal of Biological Sciences; 6(7):806-826.] | ||
| + | |||
8. [http://jcb.rupress.org/content/143/3/777/tab-pdf Ebneth A, Godemann K, Stamer S, Illenberger B, Trinczek E, et. al. 1998. “Overexpression of Tau Protein Inhibits Kinesin-dependent Trafficking of Vesicles, Mitochondria, and Endoplasmic Reticulum: Implications for Alzheimer’s Disease.” Journal of Cell Biology; 143(3):777-794.] | 8. [http://jcb.rupress.org/content/143/3/777/tab-pdf Ebneth A, Godemann K, Stamer S, Illenberger B, Trinczek E, et. al. 1998. “Overexpression of Tau Protein Inhibits Kinesin-dependent Trafficking of Vesicles, Mitochondria, and Endoplasmic Reticulum: Implications for Alzheimer’s Disease.” Journal of Cell Biology; 143(3):777-794.] | ||
| + | |||
9. [https://genomebiology.biomedcentral.com/articles/10.1186/gb-2009-10-10-242 Shepard, P. J., & Hertel, K. J. 2009. “The SR protein family.” Genome biology; 10(10):242.] | 9. [https://genomebiology.biomedcentral.com/articles/10.1186/gb-2009-10-10-242 Shepard, P. J., & Hertel, K. J. 2009. “The SR protein family.” Genome biology; 10(10):242.] | ||
| + | |||
10. [https://www.frontiersin.org/articles/10.3389/fgene.2015.00142/fullNaro C, Bielli P, Pagliarini V, and Sette C. 2015. "The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability." Frontiers in Genetics; 6(42):1-10.] | 10. [https://www.frontiersin.org/articles/10.3389/fgene.2015.00142/fullNaro C, Bielli P, Pagliarini V, and Sette C. 2015. "The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability." Frontiers in Genetics; 6(42):1-10.] | ||
<references/> | <references/> | ||
Revision as of 01:26, 3 April 2018
Contents |
Introduction
Overview
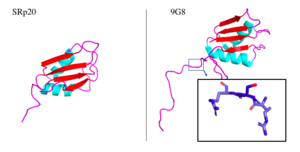
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other eukaryotes. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM).
History
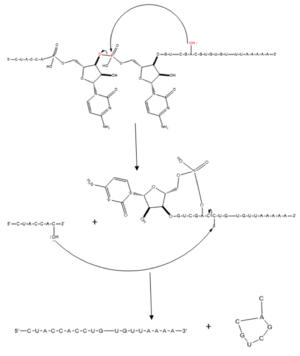
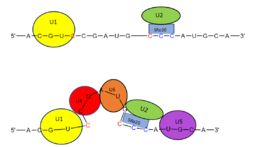
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. Alternative splicing allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins (hnRNPs). The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the SF2 (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). An identical protein, called X16, was discovered in an earlier paper studying different genes that change expression during B-cell development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013). The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006).
| |||||||||||
References
9. Shepard, P. J., & Hertel, K. J. 2009. “The SR protein family.” Genome biology; 10(10):242.