This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Isabella Gieck/Sandbox 1
From Proteopedia
| Line 7: | Line 7: | ||
HRP1 is made up of two [https://en.wikipedia.org/wiki/RNA RNA] binding domains (RBDs) that contain residues serving to facilitate RNA recognition. These two domains fold into a βαββαβ [https://en.wikipedia.org/wiki/Protein_secondary_structure secondary structure]<ref>Clery, Antoine, et al. “RNA Recognition Motifs: Boring? Not Quite.” Current Opinion in Structural Biology, Elsevier Current Trends, 30 May 2008, www.sciencedirect.com/science/article/pii/S0959440X08000584.</ref> in an RNA-free environment, allowing Hrp1 to behave rigidly. The <scene name='78/782604/First_rbd/4'>first RBD</scene> includes residues extending from Ser158 to Ala233 and the <scene name='78/782604/Second_rbd/2'>second RBD</scene> extends from Lys244 to Ala318. Both RBDs are composed of a four-stranded [https://en.wikipedia.org/wiki/Beta_sheet beta sheet] with two [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] spanning across one side of the sheet. The linker region is made up of residues spanning from Ile234 to Gly243. When RNA is introduced into the environment, conformational change is demonstrated within the linker region and a <scene name='78/782604/Linker_helix/2'>short two-turn alpha helix</scene> forms from Arg236 to Lys241. The helix that forms is made up of many charged polar residues that [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) stablilize] themselves through <scene name='78/782604/Salt_bridges/2'>salt bridge interactions</scene> between Arg236-Asp240 and Asp237-Lys241.<ref>Perez-Canadillas, Jose Manuel. “Grabbing the Message: Structural Basis of MRNA 3â²UTR Recognition by Hrp1.” The EMBO Journal, vol. 25, no. 13, 2006, pp. 3167–3178., doi:10.1038/sj.emboj.7601190. <ref name= "rna"> </ref> | HRP1 is made up of two [https://en.wikipedia.org/wiki/RNA RNA] binding domains (RBDs) that contain residues serving to facilitate RNA recognition. These two domains fold into a βαββαβ [https://en.wikipedia.org/wiki/Protein_secondary_structure secondary structure]<ref>Clery, Antoine, et al. “RNA Recognition Motifs: Boring? Not Quite.” Current Opinion in Structural Biology, Elsevier Current Trends, 30 May 2008, www.sciencedirect.com/science/article/pii/S0959440X08000584.</ref> in an RNA-free environment, allowing Hrp1 to behave rigidly. The <scene name='78/782604/First_rbd/4'>first RBD</scene> includes residues extending from Ser158 to Ala233 and the <scene name='78/782604/Second_rbd/2'>second RBD</scene> extends from Lys244 to Ala318. Both RBDs are composed of a four-stranded [https://en.wikipedia.org/wiki/Beta_sheet beta sheet] with two [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] spanning across one side of the sheet. The linker region is made up of residues spanning from Ile234 to Gly243. When RNA is introduced into the environment, conformational change is demonstrated within the linker region and a <scene name='78/782604/Linker_helix/2'>short two-turn alpha helix</scene> forms from Arg236 to Lys241. The helix that forms is made up of many charged polar residues that [https://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) stablilize] themselves through <scene name='78/782604/Salt_bridges/2'>salt bridge interactions</scene> between Arg236-Asp240 and Asp237-Lys241.<ref>Perez-Canadillas, Jose Manuel. “Grabbing the Message: Structural Basis of MRNA 3â²UTR Recognition by Hrp1.” The EMBO Journal, vol. 25, no. 13, 2006, pp. 3167–3178., doi:10.1038/sj.emboj.7601190. <ref name= "rna"> </ref> | ||
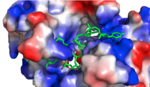
| - | [[Image:Screen Shot 2018-04-02 at 9.57.35 PM.png |150px|left|thumb|'''Figure 1:'''Positively charged cleft within HRP1 in which RNA binds | + | [[Image:Screen Shot 2018-04-02 at 9.57.35 PM.png |150px|left|thumb|'''Figure 1:'''Positively charged cleft within HRP1 in which RNA binds]] |
Hrp1 identifies the [https://en.wikipedia.org/wiki/Polyadenylation polyadenylation] enhancement element (PEE) in the 3' UTR of RNA through [https://en.wikipedia.org/wiki/Van_der_Waals_force Van Der Waals] interactions and [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding]. The PEE is made up of UAUAUA sequence in which HRP1 recognizes and binds. The association of these RBD domains aids in binding through the development of a deep highly positively charged cleft between the two. This cleft is v-shaped, accommodating the RNA very well. Some RNA bases have been revealed to be buried within protein pockets while some are observed in between protein loops. The recognition of RNA by Hrp1 is entirely dominated by RNA-protein interactions. The absence of RNA base pairing or RNA base-base stacking which have been observed in similar complexes might explain the ability of Hrp1 to target short RNA sequences in comparison to those recognized by similar proteins. | Hrp1 identifies the [https://en.wikipedia.org/wiki/Polyadenylation polyadenylation] enhancement element (PEE) in the 3' UTR of RNA through [https://en.wikipedia.org/wiki/Van_der_Waals_force Van Der Waals] interactions and [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding]. The PEE is made up of UAUAUA sequence in which HRP1 recognizes and binds. The association of these RBD domains aids in binding through the development of a deep highly positively charged cleft between the two. This cleft is v-shaped, accommodating the RNA very well. Some RNA bases have been revealed to be buried within protein pockets while some are observed in between protein loops. The recognition of RNA by Hrp1 is entirely dominated by RNA-protein interactions. The absence of RNA base pairing or RNA base-base stacking which have been observed in similar complexes might explain the ability of Hrp1 to target short RNA sequences in comparison to those recognized by similar proteins. | ||
Revision as of 02:28, 3 April 2018
Contents |
HRP1 found in Saccharomyces cerevisiae
|
Introduction
Structure
HRP1 is made up of two RNA binding domains (RBDs) that contain residues serving to facilitate RNA recognition. These two domains fold into a βαββαβ secondary structure[1] in an RNA-free environment, allowing Hrp1 to behave rigidly. The includes residues extending from Ser158 to Ala233 and the extends from Lys244 to Ala318. Both RBDs are composed of a four-stranded beta sheet with two alpha helices spanning across one side of the sheet. The linker region is made up of residues spanning from Ile234 to Gly243. When RNA is introduced into the environment, conformational change is demonstrated within the linker region and a forms from Arg236 to Lys241. The helix that forms is made up of many charged polar residues that stablilize themselves through between Arg236-Asp240 and Asp237-Lys241.[2]
Hrp1 identifies the polyadenylation enhancement element (PEE) in the 3' UTR of RNA through Van Der Waals interactions and hydrogen bonding. The PEE is made up of UAUAUA sequence in which HRP1 recognizes and binds. The association of these RBD domains aids in binding through the development of a deep highly positively charged cleft between the two. This cleft is v-shaped, accommodating the RNA very well. Some RNA bases have been revealed to be buried within protein pockets while some are observed in between protein loops. The recognition of RNA by Hrp1 is entirely dominated by RNA-protein interactions. The absence of RNA base pairing or RNA base-base stacking which have been observed in similar complexes might explain the ability of Hrp1 to target short RNA sequences in comparison to those recognized by similar proteins.
RNA binding
Recognition Specificity
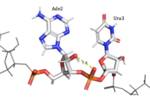
HRP1 is able to discriminate RNA from DNA due to ribose-specific hydrogen bonds, such as the ones found between the 2’OH of Ade2 and phosphate oxygen of Ura3, and between the 2’OH of Ade6 and 5’O of Ura7.[3] Furthermore, the residues found in the binding site of HRP1 have specific interactions with each individual base of the 5’-AUAUAU’3’ RNA sequence. The adenines are deeply buried in hydrophobic pockets, and uracil recognition is mainly dependent on Van der Waals interactions.
Adenosine recognition
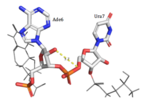
HRP1 specifically binds 3 Adenosine ribonucleotides within the PEE. Adenosine recognition is facilitated through the use of hydrophobic pockets found within HRP1. Ade2 binding is made possible through the interaction of Phe246, which makes up the foundation of the recognition pocket. In addition, the C-terminal of Arg321 interacts with the opposite side of Ade2 through pi-cation interactions. Upon binding, Ade4 is fit inside of a deep hydrophobic pocket made up of Trp168 and Lys226. The stacking of Trp in this interaction is demonstrated as a unique feature of Hrp1; similar Hrp1-like proteins maintain this conserved Trp, but do not demonstrate Trp stacking. The hydrophobic pocket in which Ade6 resides upon binding is made up of Phe162 and Ile234, which sandwich Ade6. In addition, the three Adenosines participating in binding are also recognized by Hydrogen bonds to bases that determine specificity. The three Adenosines recognized display 1, 3, and 2 Hydrogen Bond(s), respectively. The three backbone amides (Glu319 NH, Trp168 NH, Ile234 NH) hydrogen bond with nitrogen atoms of the three adenosine bases (Ade2 N1, Ade4 N7, Ade6 N1). In addition, Ade4 makes base specific contacts with Asn167 and Lys226. Ade4 acts as the donor in it’s interaction with Lys226 and as the acceptor with Asn167. Ade6 interacts with Arg232 in which it acts as the donor.[3]
Uracil recognition
Hrp1 interacts with the three uracil bases mainly though Van der Waals contacts. All three uridines interact with aromatic residues of the protein, although Ura3 and Ura5 have low surface accessibility compared to Ura7. Ura3 interacts with Phe288 in a nonplanar position, and Ura5 behaves the same with Phe162. However, Ura7 does form a planar stacking arrangement with the aromatic ring of Phe202. Base discrimination from cytosine relies heavily on the hydrogen bonding between the O4 of the uracil base and the amine group of Lys160. The O2 and O4 of Ura5 also play a role in base discrimination as they form hydrogen bonds with Lys244 and Lys231, respectively. While these two lysines are conserved in most Hrp1-like proteins, there are variations in other organisms that replace Lys244 with an Asn, although the interaction remains conserved. Discrimination at Ura3 is less clear, but it is suggested that it is due to the interaction of its N3 with the backbone phosphate of Ade6.
Novelty
While Trp168 is a highly conserved residue amongst fungal HRP1-like proteins, the tryptophan stacking found with Ade4 appears to be a novel feature not found in other single-stranded RNA and 2 RBD complexes. It is believed that Trp168 and the ring stacking it engages in are crucial to the affinity and base specificity for RNA.
To research Trp168's importance, studies have experimentally replaced it with phenylalanine and alanine side chains. While these mutants retained the ability to form 1 protein:1 RNA ratio complexes, the affinity for the AU repetition PEE is at least 10 times weaker than that of the wild-type. Furthermore, results of 15N-HSQC spectra comparisons suggest that while the Phe and Ala mutants share similar RNA-binding modes to each, they are completely different from the wild-type Trp.
Another structural finding of the splicing-factor Fox-1 in complex with RNA identifies Phe126 to have an equivalent position as Trp128 has in Hrp1. Studies were done to test the importance of Phe126 in RNA binding by mutating this residue. Similarly to the experiment mentioned above, it was concluded that the aromatic structure of Phe126 played an important role with affinity, as the residue engages in 2 planar stacking interactions with 2 RNA bases and makes contact with a third base. However, aromatic mutants did not have a significant effect on affinity, which suggests that they share a similar binding mode to the Phe126 wild-type. This is not the case with Trp168 in Hrp1, indicating that perhaps Hrp1 has strict sequence requirements.[3]
References
- ↑ Clery, Antoine, et al. “RNA Recognition Motifs: Boring? Not Quite.” Current Opinion in Structural Biology, Elsevier Current Trends, 30 May 2008, www.sciencedirect.com/science/article/pii/S0959440X08000584.
- ↑ Perez-Canadillas, Jose Manuel. “Grabbing the Message: Structural Basis of MRNA 3â²UTR Recognition by Hrp1.” The EMBO Journal, vol. 25, no. 13, 2006, pp. 3167–3178., doi:10.1038/sj.emboj.7601190. <ref> </li> <li id="cite_note-rna">[[#cite_ref-rna_2|↑]] <strong class="error">Cite error: Invalid <code><ref></code> tag; no text was provided for refs named <code>rna</code></strong></li></ol></ref>