This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Isabella Gieck/Sandbox 1
From Proteopedia
| Line 36: | Line 36: | ||
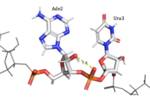
[[Image:Ade6-Ura7 H-Bonding.png|150px|right|thumb|'''Figure 3:'''Ade6 donating a hydrogen to the 5'O of Ura7.]] | [[Image:Ade6-Ura7 H-Bonding.png|150px|right|thumb|'''Figure 3:'''Ade6 donating a hydrogen to the 5'O of Ura7.]] | ||
=== Adenosine recognition === | === Adenosine recognition === | ||
| - | HRP1 specifically binds 3 Adenosine ribonucleotides within the PEE. Adenosine recognition is facilitated through the use of hydrophobic pockets found within HRP1. Ade2 binding is made possible through the interaction of Phe246, which makes up the foundation of the recognition pocket. In addition, the C-terminal of Arg321 interacts with the opposite side of Ade2 through pi-cation interactions. Upon binding, Ade4 is fit inside of a deep hydrophobic pocket made up of Trp168 and Lys226. The stacking of Trp in this interaction is demonstrated as a unique feature of Hrp1; similar Hrp1-like proteins maintain this conserved Trp, but do not demonstrate Trp stacking. The hydrophobic pocket in which Ade6 resides upon binding is made up of Phe162 and Ile234, which sandwich Ade6. In addition, the three Adenosines participating in binding are also recognized by Hydrogen bonds to bases that determine specificity. The three Adenosines recognized display 1, 3, and 2 Hydrogen Bond(s), respectively. The three backbone amides (Glu319 NH, Trp168 NH, Ile234 NH) hydrogen bond with nitrogen atoms of the three adenosine bases (Ade2 N1, Ade4 N7, Ade6 N1). In addition, Ade4 makes base specific contacts with Asn167 and Lys226. Ade4 acts as the donor in it’s interaction with Lys226 and as the acceptor with Asn167. Ade6 interacts with Arg232 in which it acts as the donor. | + | HRP1 specifically binds 3 Adenosine ribonucleotides within the PEE. Adenosine recognition is facilitated through the use of hydrophobic pockets found within HRP1. Ade2 binding is made possible through the interaction of Phe246, which makes up the foundation of the recognition pocket. In addition, the C-terminal of Arg321 interacts with the opposite side of Ade2 through pi-cation interactions. Upon binding, Ade4 is fit inside of a deep hydrophobic pocket made up of Trp168 and Lys226. The stacking of Trp in this interaction is demonstrated as a unique feature of Hrp1; similar Hrp1-like proteins maintain this conserved Trp, but do not demonstrate Trp stacking. The hydrophobic pocket in which Ade6 resides upon binding is made up of Phe162 and Ile234, which sandwich Ade6. In addition, the three Adenosines participating in binding are also recognized by Hydrogen bonds to bases that determine specificity. The three Adenosines recognized display 1, 3, and 2 Hydrogen Bond(s), respectively. The three backbone amides (Glu319 NH, Trp168 NH, Ile234 NH) hydrogen bond with nitrogen atoms of the three adenosine bases (Ade2 N1, Ade4 N7, Ade6 N1). In addition, Ade4 makes base specific contacts with Asn167 and Lys226. Ade4 acts as the donor in it’s interaction with Lys226 and as the acceptor with Asn167. Ade6 interacts with Arg232 in which it acts as the donor. |
<scene name='78/782604/Adenosine_2_interactions/1'>Adenosine 2 interactions</scene> | <scene name='78/782604/Adenosine_2_interactions/1'>Adenosine 2 interactions</scene> | ||
| Line 56: | Line 56: | ||
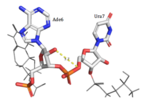
[[Image:Screen Shot 2018-04-02 at 10.13.18 PM.png |150px|left|thumb|'''Figure 4:'''Trp168 "stacking" onto Ade4 through Van Der Waals interactions.]] | [[Image:Screen Shot 2018-04-02 at 10.13.18 PM.png |150px|left|thumb|'''Figure 4:'''Trp168 "stacking" onto Ade4 through Van Der Waals interactions.]] | ||
| - | Another structural finding of the splicing-factor [https://www.rcsb.org/structure/4zka Fox-1] in complex with RNA identifies Phe126 to have an equivalent position as Trp128 has in Hrp1. Studies were done to test the importance of Phe126 in RNA binding by mutating this residue. Similarly to the experiment mentioned above, it was concluded that the aromatic structure of Phe126 played an important role with affinity, as the residue engages in 2 planar stacking interactions with 2 RNA bases and makes contact with a third base. However, aromatic mutants did not have a significant effect on affinity, which suggests that they share a similar binding mode to the Phe126 wild-type. This is not the case with Trp168 in Hrp1, indicating that perhaps Hrp1 has strict sequence requirements. | + | Another structural finding of the splicing-factor [https://www.rcsb.org/structure/4zka Fox-1] in complex with RNA identifies Phe126 to have an equivalent position as Trp128 has in Hrp1. Studies were done to test the importance of Phe126 in RNA binding by mutating this residue. Similarly to the experiment mentioned above, it was concluded that the aromatic structure of Phe126 played an important role with affinity, as the residue engages in 2 planar stacking interactions with 2 RNA bases and makes contact with a third base. However, aromatic mutants did not have a significant effect on affinity, which suggests that they share a similar binding mode to the Phe126 wild-type. This is not the case with Trp168 in Hrp1, indicating that perhaps Hrp1 has strict sequence requirements. |
== Disease == | == Disease == | ||
| - | Though Hrp1 is not analogous to any mammalian hnRNP<ref> Gross, S., and C. Moore. “Five Subunits Are Required for Reconstitution of the Cleavage and Polyadenylation Activities of Saccharomyces Cerevisiae Cleavage Factor I.” Proceedings of the National Academy of Sciences, vol. 98, no. 11, Aug. 2001, pp. 6080–6085., doi:10.1073/pnas.101046598. </ref>, the protein and its corresponding gene are occasionally studied as orthologues to human hnRNPs. HNRPDL is one such family of human hnRNPs. Mutations to several members of this class of hnRNPs result in many facets of muscular dystrophy. A study by Vieira, et al.<ref> Vieira, Natássia M., et al. “A Defect in the RNA-Processing Protein HNRPDL Causes Limb-Girdle Muscular Dystrophy 1G (LGMD1G).” Human Molecular Genetics, vol. 23, no. 15, 2014, pp. 4103–4110., doi:10.1093/hmg/ddu127. </ref> found that elimination of Hrp1 had profound effects on protein localization and activation, and these results were used as a model for the genotypic causation of muscular dystrophy. | + | Though Hrp1 is not analogous to any mammalian hnRNP<ref>Gross, S., and C. Moore. “Five Subunits Are Required for Reconstitution of the Cleavage and Polyadenylation Activities of Saccharomyces Cerevisiae Cleavage Factor I.” Proceedings of the National Academy of Sciences, vol. 98, no. 11, Aug. 2001, pp. 6080–6085., doi:10.1073/pnas.101046598.</ref>, the protein and its corresponding gene are occasionally studied as orthologues to human hnRNPs. HNRPDL is one such family of human hnRNPs. Mutations to several members of this class of hnRNPs result in many facets of muscular dystrophy. A study by Vieira, et al. <ref> Vieira, Natássia M., et al. “A Defect in the RNA-Processing Protein HNRPDL Causes Limb-Girdle Muscular Dystrophy 1G (LGMD1G).” Human Molecular Genetics, vol. 23, no. 15, 2014, pp. 4103–4110., doi:10.1093/hmg/ddu127.</ref> found that elimination of Hrp1 had profound effects on protein localization and activation, and these results were used as a model for the genotypic causation of muscular dystrophy. |
| + | |||
== References == | == References == | ||
<references /> | <references /> | ||
Revision as of 03:31, 3 April 2018
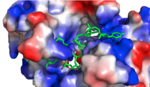
HRP1 found in Saccharomyces cerevisiae
Introduction
Hrp1 is a heterogeneous ribonuclear protein of Saccharomyces cerevisiae, baker’s yeast. Hrp1 is an essential component of 3’ pre-mRNA processing and contributes to the preparatory cleavage required for polyadenylation. The gene expressed as Hrp1, HRP1, was first isolated by Henry, et al.[1] and was later attributed to the Hrp1 protein by Kessler, et al.[2] Hrp1 also participates in the regulation of the 3’ end.
| |||||||||||