User:David Ryskamp/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). It is comprised of four [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motifs] (RRMs), which are highly conserved RNA-binding domains.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> The RRM in PABP is found in over two hundred families of proteins across species, indicating that it is ancient.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> RRM1 and 2 are examined in this article. Each RRM has a four-stranded antiparallel beta sheet backed by two corresponding alpha helices. <ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> mRNA poly-adenosine recognition is due to the presence of the conserved residues within the beta-sheet surface <ref name="The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.">Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.</ref> , which forms a <scene name='78/782616/Trough2/1'>trough</scene>-like pocket for the mRNA to bind. The beta-sheet flooring present in PABP interacts with the 3’ mRNA tail via a combination of van der Waals, aromatic stacking, and Hydrogen bonding. Three major aromatic stacking interactions are <scene name='78/782616/A3-phe102/1'>Phe102 with A3</scene>, <scene name='78/782616/Tyr56-a8/1'>Tyr56 with A8</scene>, and <scene name='78/782616/A6-tyr14/1'>Tyr14 with A6</scene>. Through these interactions, PABP binds to 3’ Poly (A) tail with a KD of 2-7 nM. <ref name="Roles of Cytoplasmic Poly(A)-Binding Proteins">Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.</ref> | The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). It is comprised of four [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motifs] (RRMs), which are highly conserved RNA-binding domains.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> The RRM in PABP is found in over two hundred families of proteins across species, indicating that it is ancient.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> RRM1 and 2 are examined in this article. Each RRM has a four-stranded antiparallel beta sheet backed by two corresponding alpha helices. <ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> mRNA poly-adenosine recognition is due to the presence of the conserved residues within the beta-sheet surface <ref name="The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.">Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.</ref> , which forms a <scene name='78/782616/Trough2/1'>trough</scene>-like pocket for the mRNA to bind. The beta-sheet flooring present in PABP interacts with the 3’ mRNA tail via a combination of van der Waals, aromatic stacking, and Hydrogen bonding. Three major aromatic stacking interactions are <scene name='78/782616/A3-phe102/1'>Phe102 with A3</scene>, <scene name='78/782616/Tyr56-a8/1'>Tyr56 with A8</scene>, and <scene name='78/782616/A6-tyr14/1'>Tyr14 with A6</scene>. Through these interactions, PABP binds to 3’ Poly (A) tail with a KD of 2-7 nM. <ref name="Roles of Cytoplasmic Poly(A)-Binding Proteins">Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.</ref> | ||
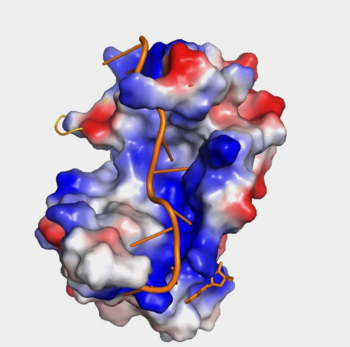
| - | Further, the RRM1/2 complex interacts with the mRNA's sugar-phosphate backbone, where 4 of the 8 mRNA adenosines interact electrostatically.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> Upon closer examination of the PABP structure, the protein contains loop-like domains that form the walls of the beta-sheet trough. Although these <scene name='78/782616/Walls_of_trough/3'>loop walls</scene> are present, no interaction occurs between the mRNA and these regions. We propose that these loops only keep unwanted cellular elements out of the binding pocket via hydrophobic and hydrophilic interactions, maintaining the protein's selectivity for mRNA. The structural elements highlighted consist of the RRM1/2 subunits, the linker domain, and the Poly(A) mRNA binding trough. | + | Further, the RRM1/2 complex interacts with the mRNA's sugar-phosphate backbone, where 4 of the 8 mRNA adenosines interact electrostatically.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> Upon closer examination of the PABP structure, the protein contains loop-like domains that form the walls of the beta-sheet trough. Although these <scene name='78/782616/Walls_of_trough/3'>loop walls</scene> are present, no interaction occurs between the mRNA and these regions. We propose that these loops only keep unwanted cellular elements out of the binding pocket via hydrophobic and hydrophilic interactions, maintaining the protein's selectivity for mRNA (Figure 1). The structural elements highlighted consist of the RRM1/2 subunits, the linker domain, and the Poly(A) mRNA binding trough. |
| + | [[Image:Hydrophobicity (1).png|350px|left|thumb| "Figure 1:" Surface hydrophobicity shown in presence of mRNA]] | ||
===RNA Recognition Motifs (RRMs)=== | ===RNA Recognition Motifs (RRMs)=== | ||
| Line 44: | Line 45: | ||
===Recognition of the Poly(A) Tail=== | ===Recognition of the Poly(A) Tail=== | ||
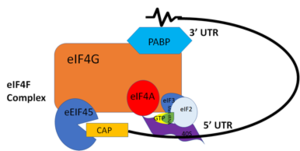
Polyadenylation of an mRNA involves the recognition of the 5’-AAUAAA-3’ consensus site, the cleavage downstream of the consensus site, and then the addition of adenines by [https://en.wikipedia.org/wiki/Polynucleotide_adenylyltransferase Poly(A) Polymerase] to the 3’ end. The newly added poly(A) tail is associated with the PABP. PABP requires 11-12 adenosines in order to bind. PABP and the bound Poly(A) tail work together to stabilize mRNA by preventing exo-ribonucleolytic degradation,<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> thereby guiding the mRNA molecule into the translation pathway. Upon mRNA poly(A) recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor [https://en.wikipedia.org/wiki/EIF4G eIF4G]. | Polyadenylation of an mRNA involves the recognition of the 5’-AAUAAA-3’ consensus site, the cleavage downstream of the consensus site, and then the addition of adenines by [https://en.wikipedia.org/wiki/Polynucleotide_adenylyltransferase Poly(A) Polymerase] to the 3’ end. The newly added poly(A) tail is associated with the PABP. PABP requires 11-12 adenosines in order to bind. PABP and the bound Poly(A) tail work together to stabilize mRNA by preventing exo-ribonucleolytic degradation,<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> thereby guiding the mRNA molecule into the translation pathway. Upon mRNA poly(A) recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor [https://en.wikipedia.org/wiki/EIF4G eIF4G]. | ||
| - | |||
| - | [[Image:Hydrophobicity (1).png|350px|left|thumb| "Figure 1:" Surface hydrophobicity shown in presence of mRNA]] | ||
===mRNA Stabilization=== | ===mRNA Stabilization=== | ||
Revision as of 17:26, 6 April 2018
Human Poly(A) Binding Protein (1CVJ)
| |||||||||||
References
- ↑ Blobel, Gunter. “A Protein of Molecular Weight 78,000 Bound to the Polyadenylate Region of Eukaryotic Messenger Rnas.” Proceedings of the National Academy of Sciences of the United States of America, vol. 70, no. 3, 1973, pp. 924–8.
- ↑ Baer, Bradford W. and Kornberg, Roger D. "The Protein Responsible for the Repeating Structure of Cytoplasmic Poly(A)-Ribonucleoprotein." The Journal of Cell Biology, vol. 96, no. 3, Mar. 1983, pp. 717-721. EBSCOhost.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print.
- ↑ Kühn, Uwe and Elmar, Wahle. “Structure and Function of Poly(a) Binding Proteins.” Bba - Gene Structure & Expression, vol. 1678, no. 2/3, 2004.
- ↑ 5.0 5.1 5.2 5.3 5.4 Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.
- ↑ Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.
- ↑ 7.0 7.1 7.2 7.3 “Oculopharyngeal Muscular Dystrophy.” NORD (National Organization for Rare Disorders), rarediseases.org/rare-diseases/oculopharyngeal-muscular-dystrophy/.
- ↑ Richard, Pascale, et al. “Correlation between PABPN1 Genotype and Disease Severity in Oculopharyngeal Muscular Dystrophy.” Neurology, vol. 88, no. 4, 2016, pp. 359–365., doi:10.1212/wnl.0000000000003554.