We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:David Ryskamp/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Background == | == Background == | ||
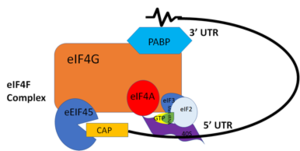
| - | The Human Poly(A) Binding Protein [https://www.rcsb.org/structure/1cvj (PABP)] was discovered in 1973 by the use of a sedimentation profile detailing the RNase digestion differentiated the PABP protein. <ref name="A Protein of Molecular Weight 78,000">Blobel, Gunter. “A Protein of Molecular Weight 78,000 Bound to the Polyadenylate Region of Eukaryotic Messenger Rnas.” Proceedings of the National Academy of Sciences of the United States of America, vol. 70, no. 3, 1973, pp. 924–8.</ref> Attempts to purify the 75 kDa protein then followed. In 1983, then considered “poly(A)-organizing protein,” was determined and purified by molecular weight, ligand-binding affinity, and amounts found in cytoplasmic portions of cell with ability to bind to free poly(A). <ref name="Cytoplasmic Poly(A)">Baer, Bradford W. and Kornberg, Roger D. "The Protein Responsible for the Repeating Structure of Cytoplasmic Poly(A)-Ribonucleoprotein." The Journal of Cell Biology, vol. 96, no. 3, Mar. 1983, pp. 717-721. EBSCOhost | + | The Human Poly(A) Binding Protein [https://www.rcsb.org/structure/1cvj (PABP)] was discovered in 1973 by the use of a sedimentation profile detailing the RNase digestion differentiated the PABP protein. <ref name="A Protein of Molecular Weight 78,000">Blobel, Gunter. “A Protein of Molecular Weight 78,000 Bound to the Polyadenylate Region of Eukaryotic Messenger Rnas.” Proceedings of the National Academy of Sciences of the United States of America, vol. 70, no. 3, 1973, pp. 924–8.</ref> Attempts to purify the 75 kDa protein then followed. In 1983, then considered “poly(A)-organizing protein,” was determined and purified by molecular weight, ligand-binding affinity, and amounts found in cytoplasmic portions of cell with ability to bind to free poly(A). <ref name="Cytoplasmic Poly(A)">Baer, Bradford W. and Kornberg, Roger D. "The Protein Responsible for the Repeating Structure of Cytoplasmic Poly(A)-Ribonucleoprotein." The Journal of Cell Biology, vol. 96, no. 3, Mar. 1983, pp. 717-721. EBSCOhost. </ref> |
| Line 11: | Line 11: | ||
== Structure == | == Structure == | ||
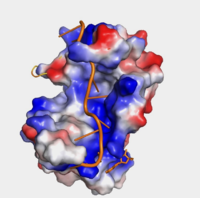
| - | The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). The subunits of PABP, RRM1 and RRM2, are examined in this article as the ''in vivo'' form seen in biological assembly 1 (via PDB). | + | The crystal structure PABP was derived from X-ray Diffraction at 2.6Å (R-value: 23%). The subunits of PABP, RRM1 and RRM2, are examined in this article as the ''in vivo'' form seen in biological assembly 1 (via PDB). The protein has a homopolymeric structure, containing four RNA recognition motifs (RRMs), which are conserved. <ref name="Structure and Function">Kühn, Uwe and Elmar, Wahle. “Structure and Function of Poly(a) Binding Proteins.” Bba - Gene Structure & Expression, vol. 1678, no. 2/3, 2004. </ref> <scene name='78/782616/Rrm1_only/2'>RRM1</scene> and <scene name='78/782616/Rrm2_only/2'>RRM2</scene> are N-terminal domains that are connected by a <scene name='78/782616/Linker/3'>linker</scene>.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> Opposed to their counterparts, RRM3 and RRM4 bind Poly (A) RNA less tightly than RRM1 and RRM2.<ref name="Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein">Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print. </ref> |
===RNA Recognition Motifs (RRMs)=== | ===RNA Recognition Motifs (RRMs)=== | ||
| Line 22: | Line 22: | ||
==Interactions== | ==Interactions== | ||
| - | ===Adenosine Recognition Interactions | + | ===Adenosine Recognition Interactions=== |
<table><tr><td colspan='2'> | <table><tr><td colspan='2'> | ||
<tr id='Nucleotide'><td class="sblockLbl"><b>Nucleotide</b></td><td class="sblockDat">PABP residue</td><td class="sblockDat">Atom on PABP residue</td> | <tr id='Nucleotide'><td class="sblockLbl"><b>Nucleotide</b></td><td class="sblockDat">PABP residue</td><td class="sblockDat">Atom on PABP residue</td> | ||
Revision as of 18:45, 6 April 2018
Human Poly(A) Binding Protein (1CVJ)
| |||||||||||
References
- ↑ Blobel, Gunter. “A Protein of Molecular Weight 78,000 Bound to the Polyadenylate Region of Eukaryotic Messenger Rnas.” Proceedings of the National Academy of Sciences of the United States of America, vol. 70, no. 3, 1973, pp. 924–8.
- ↑ Baer, Bradford W. and Kornberg, Roger D. "The Protein Responsible for the Repeating Structure of Cytoplasmic Poly(A)-Ribonucleoprotein." The Journal of Cell Biology, vol. 96, no. 3, Mar. 1983, pp. 717-721. EBSCOhost.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print.
- ↑ 4.0 4.1 4.2 4.3 4.4 Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.
- ↑ Kühn, Uwe and Elmar, Wahle. “Structure and Function of Poly(a) Binding Proteins.” Bba - Gene Structure & Expression, vol. 1678, no. 2/3, 2004.
- ↑ Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.
- ↑ 7.0 7.1 7.2 7.3 “Oculopharyngeal Muscular Dystrophy.” NORD (National Organization for Rare Disorders), rarediseases.org/rare-diseases/oculopharyngeal-muscular-dystrophy/.
- ↑ Richard, Pascale, et al. “Correlation between PABPN1 Genotype and Disease Severity in Oculopharyngeal Muscular Dystrophy.” Neurology, vol. 88, no. 4, 2016, pp. 359–365., doi:10.1212/wnl.0000000000003554.