2gr9
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 2gr9 |SIZE=350|CAPTION= <scene name='initialview01'>2gr9</scene>, resolution 3.1Å | |PDB= 2gr9 |SIZE=350|CAPTION= <scene name='initialview01'>2gr9</scene>, resolution 3.1Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=GLU:GLUTAMIC+ACID'>GLU</scene>, <scene name='pdbligand=NAD:NICOTINAMIDE-ADENINE-DINUCLEOTIDE'>NAD</scene> |
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Pyrroline-5-carboxylate_reductase Pyrroline-5-carboxylate reductase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.5.1.2 1.5.1.2] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Pyrroline-5-carboxylate_reductase Pyrroline-5-carboxylate reductase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.5.1.2 1.5.1.2] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY=[[2gra|2GRA]] | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2gr9 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2gr9 OCA], [http://www.ebi.ac.uk/pdbsum/2gr9 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=2gr9 RCSB]</span> | ||
}} | }} | ||
| Line 27: | Line 30: | ||
[[Category: Meng, Z.]] | [[Category: Meng, Z.]] | ||
[[Category: Rao, Z.]] | [[Category: Rao, Z.]] | ||
| - | + | [[Category: crystal strucutre,human pyrroline-5-carboxylate reductase]] | |
| - | + | ||
| - | [[Category: crystal strucutre | + | |
| - | + | ||
[[Category: nadh]] | [[Category: nadh]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 03:20:23 2008'' |
Revision as of 00:20, 31 March 2008
| |||||||
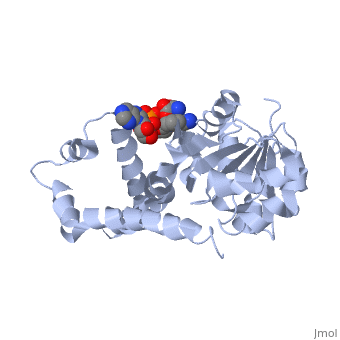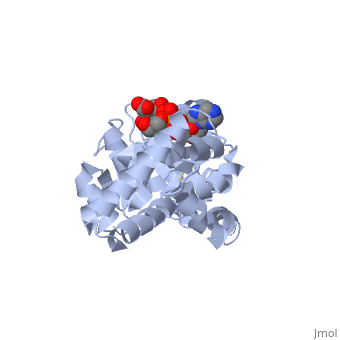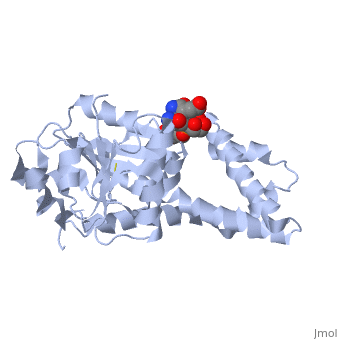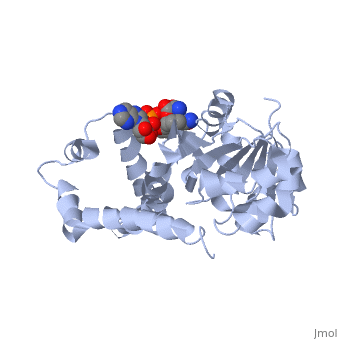
| , resolution 3.1Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Activity: | Pyrroline-5-carboxylate reductase, with EC number 1.5.1.2 | ||||||
| Related: | 2GRA
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Crystal structure of P5CR complexed with NADH
Overview
Pyrroline-5-carboxylate reductase (P5CR) is a universal housekeeping enzyme that catalyzes the reduction of Delta(1)-pyrroline-5-carboxylate (P5C) to proline using NAD(P)H as the cofactor. The enzymatic cycle between P5C and proline is very important for the regulation of amino acid metabolism, intracellular redox potential, and apoptosis. Here, we present the 2.8 Angstroms resolution structure of the P5CR apo enzyme, its 3.1 Angstroms resolution ternary complex with NAD(P)H and substrate-analog. The refined structures demonstrate a decameric architecture with five homodimer subunits and ten catalytic sites arranged around a peripheral circular groove. Mutagenesis and kinetic studies reveal the pivotal roles of the dinucleotide-binding Rossmann motif and residue Glu221 in the human enzyme. Human P5CR is thermostable and the crystals were grown at 37 degrees C. The enzyme is implicated in oxidation of the anti-tumor drug thioproline.
About this Structure
2GR9 is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Crystal structure of human pyrroline-5-carboxylate reductase., Meng Z, Lou Z, Liu Z, Li M, Zhao X, Bartlam M, Rao Z, J Mol Biol. 2006 Jun 23;359(5):1364-77. Epub 2006 May 11. PMID:16730026
Page seeded by OCA on Mon Mar 31 03:20:23 2008