Background
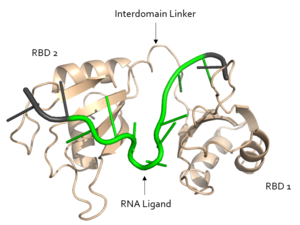
Figure 1. Structural overview of Sxl. RNA ligand colored in green is recognized and bound, RNA ligand colored in grey is not bound. Structure shown is
PDB:1b7f. Figure created in PyMol.
Sex Lethal Protein (Sxl) is a splicing repressor of the male developmental pathway of sex determination of the common fruit fly, Drosophila melanogaster[1]. Sxl regulates alternative splicing pathways to promote the expression of female sex-linked proteins. In eukaryotes, splicing is carried out via the spliceosome, a ribozyme-protein complex which binds to the 5’ and 3’ splice sites. As Sxl is a splicing repressor, it prevents the binding of the U2AF and U1 subunits of the spliceosome at their respective splice sites, which represses the alternative splicing mechanism[2]. As a result, the fruit fly expressing Sxl will produce mRNA transcripts encoding proteins for the female developmental pathway[1]. If Sxl is unable to repress translation of the male-sex lethal (msl-2) protein in female flies, the female fly will die due to hyperexpression of both X chromosomes[3][4].
The Sxl RNA splicing targets encode for the transformer (tra) and msl-2 proteins. Tra is a splicing activator for the female developmental pathway, and msl-2 expression modulates X chromosome application in male fruit flies. The mechanism for how Sxl targets these pathways differs slightly. In both mechanisms, Sxl occupies the 3' splice site and prevents U2AF from binding. This causes the U2AF splicing factor to bind at a downstream splice site encoding proteins in the female developmental pathway. In msl-2 targeting, Sxl also blocks the binding of another regulatory splicing factor, Rox8, and the U1 snRNP at the 5’ splice site[2]. Sxl can also control its own splicing pattern to conserve female expression. It does so by binding to Exon 3 of its own RNA and creating an RNP complex to eliminate this exon. After removal of Exon 3, Sxl becomes active and female expression is maintained.
Structure
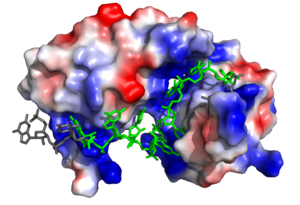
Figure 2. Sex-lethal protein showing the electropositive binding pocket and the bound RNA ligand. Pre-mRNA residues binding to Sxl shown in green, non-binding residues shown in grey. Structure shown is
PDB:1b7f. Figure created in PyMol.
Sxl is composed of two asymmetric RNA binding domains (RBD1 and RBD2) which recognize a poly-uridine site in the pre-mRNA transcript[1]. Each RBD is comprised of two alpha helices and one antiparallel four-stranded β sheet[1] containing the RNA recognition motif(Fig. 1). The β sheets face each other, lining the V-shaped cleft[1], shown in sand in Fig. 1. The inter-domain linker, shown in sand in Fig. 1, forms a distorted 310 helix which helps form the V-shaped cleft into which the pre-mRNA sequence binds[1][3]. Sxl binds to UGUUUUUUU sequence of GUUGUUUUUUUU in the tra pre-mRNA[1][3]. RBD1 binds U6-U11 and RBD2 binds U3, G4, and U5. Figure 1 shows bound pre-mRNA residues in green and non-bound pre-mRNA residues in grey. Although the two RBDs do not interact with each other, this nine-ribonucleotide sequence must be recognized continuously to allow Sxl to bind, preventing U2AF from binding at the 3’ splice site[1]. The binding of Sxl to the pre-mRNA occurs in an electropositive pocket (shown in blue in Fig. 2) due to extensive interactions with the RNA phosphate backbone and negatively charged residues[1]. Since Sxl binds primarily with the phosphate backbone, the protein residues are not highly conserved.
Structural Basis for Recognition of Poly-U Sequences
The structural interactions with regards to the targeting of the 5' splice site and of its own mRNA transcript are much less understood than the competition of with U2AF at the 3' splice site. All the RNA-protein interactions described here refer to tra pre-mRNA-Sxl interactions.
The is crucial to pre-mRNA binding; a mutation of R252 to alanine eliminated the ability of Sxl to bind RNA[1].
The ligand pre-mRNA sequence forms a at U5, U6, and U7. This interaction is stabilized by π stacking between the G4 and as well as U5 and , respectively[1]. The nucleobases are exposed to residues on Sxl due to the 2’ endo conformation of all the nucleotides except for U8, which maintains a 3’ endo conformation[1].
The U6 residue is recognized as part of the RNA by R195. The R195 amide hydrogen-bonds to the O2' of U6 and the U6 N3H hydrogen bonds to the R195 carbonyl oxygen[1].
In the RNA , the U7 and U8 bases are involved in π , stabilizing the 3' endo conformation of the U8 sugar[1]. U8 is further stabilized via hydrogen bonding [1]. The amine group of U8 hydrogen bonds to the the carbonyl oxygens of both S165 and Y166 [1].
is recognized by the interdomain linker [1]. This interaction is a salt bridge between the N130 side chain and a phosphate oxygen of U9[1]. U9 is further stabilized by a second between the U9 O2' and the side chain of R202 and the U9 O4' and the K197 side chain[1].
U9 facilitates the stabilization of U10, which is also recognized by the interdomain linker. to form a salt bridge.
U11 is recognized by R155. The O2' of U11 to form a hydrogen bond[1].
The above interactions are relevant in that Sxl recognizes the specific pre-mRNA based mostly on interactions with the sugar-phosphate backbones[1]. Many proteins with RNA recognition motifs are specific in the interactions they form with the bases of the RNA recognized. In contrast, Sxl has a high specificity despite primarily interacting with the phosphate backbone.
Alternative Splicing Pathways
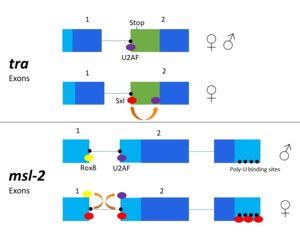
Figure 3. 2-dimensional representation of alternative splicing repression by Sxl on the
tra and
msl-2 genes.
The alternative splicing pathways of Sxl differ, but both involve repression at the 3' splice site[1][3]. The tra expression pathway only involves the 3' splice site, while the msl-2 pathway involves both the 3' splice site and the 5' splice site. Both mechanisms cause U2AF binding downstream with lower affinity (Fig. 3)[3]. U2AF is a more general splicing factor than Sxl, and prefers cytidine-containing poly-uridine pre-mRNA sequences, so Sxl binds to the guanosine-containing pre-mRNA with a 104-fold greater affinity[1].
Autoregulation
Sxl is capable of autoregulation of its expression[3]. Sxl protein autoregulates negatively by binding the poly-U sequence in its own 5'UTR, repressing translation[3]. The negative autoregulation results in maintaining appropriate concentrations of Sxl. Sxl can also positively autoregulate its expression: the Sxl gene is transcribed in male flies, but the inclusion of exon 3 results in a premature stop codon, producing an inactive, truncated protein[3]. The same Sxl promoter is active in female flies, but an additional (briefly active) Sxl promoter produces a transcript with exon 3 removed, resulting in an active Sxl protein which will initiate other female-specific splicing cascades[3].
Tra
In alternative splicing of the tra gene, Sxl binds at the 3' poly-uridine site (Fig. 3). This causes U2AF to bind downstream and the ribosome assembles at the following exon[2]. In the absence of Sxl, the normal gene for male development is transcribed. The exon contains a stop codon which results in a truncated, non-functional protein[3]. In the presence of Sxl, this exon is spliced, so the stop codon is skipped[3] (Fig. 3). This enables translation of an active tra protein[3], which is vital to female fruit fly development.
Msl-2
Msl-2 is responsible for dosage compensation of X chromosomes in fruit flies. The alternative splicing of msl-2 is reliant on Sxl binding to both the 5' and 3' splice sites (Fig. 3). Sxl binds at the 3' splice site, replacing U2AF as in tra splicing. Sxl also competes with Rox8, which binds to the first intron. As a result, Sxl prevents splicing of the first intron of the msl-2 primary transcript[4]. Sxl also binds to the poly- U sequences of the 3' UTR to repress translation (Fig.3)[2], leading to female expression. When Sxl targets msl-2, the first intron is retained[3][4]. However, the retained intron is in the 5' UTR and does not affect the reading frame[3]. When msl-2 is expressed, the X chromosome's transcription is repressed. In male fruit flies, msl-2 must be inactivated to allow increased X chromosome transcription[4]. As the expression of msl-2 is exclusively required for male fruit fly development, any mutation in Sxl which causes splicing and activation of the msl-2 gene in females leads to female fly death by hyperexpression of both X chromosomes[4].
Relevance
As Sxl functions as a splicing repressor, it may give insight into the effects of varying mechanisms of alternate splicing both in flies and other species. Sxl may also lead to understanding of human alternative splicing factors. As an RNA binding protein, research regarding Sxl may contribute to the understanding of enzymes with RNA recognition motifs. The Sxl RNP motif of RBD1 is also conserved in the ELAV family of proteins[1]. Sxl is only one of many proteins which regulate dosage compensation, but is one that is observed in XX/XY systems, and thus could lead to better comprehension of dosage compensation in species with similar sex determination systems.



