We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox GGC14
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
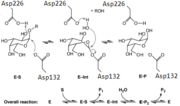
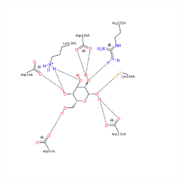
| - | This <scene name='78/781216/1t0o_alpha-galactosidase/3'>view</scene> shows the carbohydrate Galactose in the pocket of the active site. The <scene name='78/781216/1t0o_alpha-galactosidase/ | + | This <scene name='78/781216/1t0o_alpha-galactosidase/3'>view</scene> shows the carbohydrate Galactose in the pocket of the active site. The <scene name='78/781216/1t0o_alpha-galactosidase/11'>two catalytic residues</scene>, Asp132 and Asp226, can be seen sandwiching the inhibitor and product of this enzyme. |
It is also interesting to note that all 5 hydroxyl groups participate in hydrogen bonding with enzyme residues. Only 5 residues are shown in <scene name='78/781216/1t0o_alpha-galactosidase/7'>this view</scene>, but there are more. | It is also interesting to note that all 5 hydroxyl groups participate in hydrogen bonding with enzyme residues. Only 5 residues are shown in <scene name='78/781216/1t0o_alpha-galactosidase/7'>this view</scene>, but there are more. | ||
Revision as of 12:45, 23 April 2018
1T0O - a-Galactosidase from Trichoderma reesei and Complex with Galactose
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Guce AI, Clark NE, Salgado EN, Ivanen DR, Kulminskaya AA, Brumer H 3rd, Garman SC. Catalytic mechanism of human alpha-galactosidase. J Biol Chem. 2010 Feb 5;285(6):3625-32. Epub 2009 Nov 25. PMID:19940122 doi:10.1074/jbc.M109.060145
- ↑ Golubev AM, Nagem RA, Brandao Neto JR, Neustroev KN, Eneyskaya EV, Kulminskaya AA, Shabalin KA, Savel'ev AN, Polikarpov I. Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism. J Mol Biol. 2004 May 28;339(2):413-22. PMID:15136043 doi:http://dx.doi.org/10.1016/j.jmb.2004.03.062