User:Peggy Skerratt/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Regulation == | == Regulation == | ||
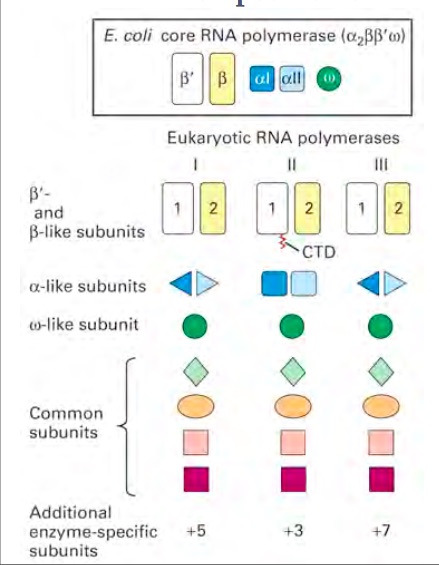
Though regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site. | Though regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site. | ||
| - | == Cancer == | + | == Connections with Cancer == |
RNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur. | RNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur. | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 22:13, 25 April 2018
[[Image:]]==RNA Polymerase==
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644