User:Peggy Skerratt/Sandbox 1
From Proteopedia
| Line 4: | Line 4: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
==Overview== | ==Overview== | ||
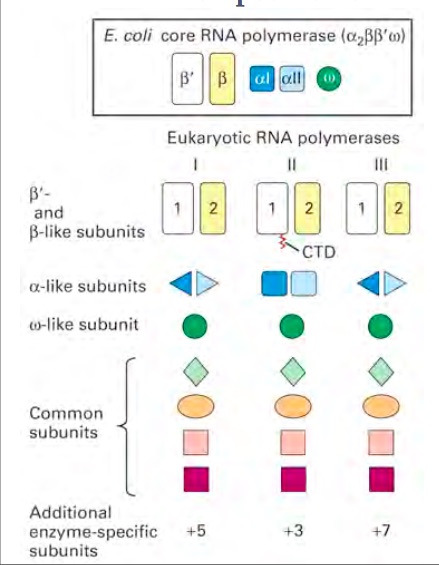
| - | RNA polymerase is an enzyme that binds to double stranded DNA and synthesizes RNA. Since the enzyme moves along the DNA strand to synthesise it, it is considered a molecular machine. RNA polymerase is similar to the DNA polymerase because it reads the DNA strand from the 3’ end to the 5’ end. It synthesizes the RNA from the 5’ end to the 3’ end. It only reads the template strand to synthesize the complementary strand. Unlike the DNA polymerase it does not need a primer to begin, it can initiate synthesis itself. | + | RNA polymerase is an enzyme that binds to double stranded DNA and synthesizes RNA. Since the enzyme moves along the DNA strand to synthesise it, it is considered a molecular machine. RNA polymerase is similar to the DNA polymerase because it reads the DNA strand from the 3’ end to the 5’ end. It synthesizes the RNA from the 5’ end to the 3’ end. It only reads the template strand to synthesize the complementary strand. Unlike the DNA polymerase it does not need a primer to begin, it can initiate synthesis itself. <ref:1>This enzyme is found in both prokaryotes and eukaryotes. There are several different types of RNA polymerases. In Prokaryotes there is only one RNA polymerase, which synthesises all of the RNA. In eukaryotes there are three polymerases (RNA polymerase I,II, and III) that synthesize different types of RNA. Prokaryote and eukaryote polymerases have similar structures. Within the eukaryotic polymerases, the enzymes are structurally similar but have different subunits attached to the polymerase. The whole process of making RNA from DNA is called transcription. Transcription has four stages: assembly, initiation, elongation and termination, in which RNA polymerase is involved. The polymerase that is found to be a key polymerase is polymerase II. The exact mechanism for the different polymerases is still being studied. Since bacteria only have one polymerase that produce RNA, it is often studied to try to understand how this process works.The process of transcription is also regulated. It is controlled through specificity factors, repressors, and activators. If the transcription factors are not regulated, oncogenic activity can occur. Overall, RNA polymerase is still being studied and research is still being conducted to uncover all of the ins and outs of RNA polymerase’s role in transcription. |
| Line 42: | Line 42: | ||
== Regulation == | == Regulation == | ||
| - | Though regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site | + | Though regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site. |
== Connections with Cancer == | == Connections with Cancer == | ||
| - | RNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur | + | RNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | < | + | <ref:1>Pratt, Charlotte, and Kathleen Cornely. “178-RNA Polymerase Structure.” YouTube, YouTube, 12 June 2014, www.youtube.com/watch?v=gqEwq4HnRmU. |
| + | Hurwitz, Jerard. “The Discovery of RNA Polymerase.” Journal of Biological Chemistry, 30 Dec. 2005, www.jbc.org/content/280/52/42477. | ||
| + | Fredericks , Mike. “Prokaryotic vs Eukaryotic Transcription.” PROKARYOTIC VS EUKARYOTIC, 2006, www.chem.uwec.edu/webpapers2006/sites/fredermr/comp.html. | ||
| + | Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. Transcription and RNA polymerase. Available from: https://www.ncbi.nlm.nih.gov/books/NBK22085/ | ||
| + | Alber , T, et al. General Biochemistry and Molecular Biology . 22 Aug. 2008, mcb.berkeley.edu/courses/mcb110/ZHOU/lec.5-7.euk_trxn_apparatus.pdf | ||
Revision as of 22:57, 25 April 2018
[[Image:]]==RNA Polymerase==
| |||||||||||
References
<ref:1>Pratt, Charlotte, and Kathleen Cornely. “178-RNA Polymerase Structure.” YouTube, YouTube, 12 June 2014, www.youtube.com/watch?v=gqEwq4HnRmU. Hurwitz, Jerard. “The Discovery of RNA Polymerase.” Journal of Biological Chemistry, 30 Dec. 2005, www.jbc.org/content/280/52/42477. Fredericks , Mike. “Prokaryotic vs Eukaryotic Transcription.” PROKARYOTIC VS EUKARYOTIC, 2006, www.chem.uwec.edu/webpapers2006/sites/fredermr/comp.html. Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. Transcription and RNA polymerase. Available from: https://www.ncbi.nlm.nih.gov/books/NBK22085/ Alber , T, et al. General Biochemistry and Molecular Biology . 22 Aug. 2008, mcb.berkeley.edu/courses/mcb110/ZHOU/lec.5-7.euk_trxn_apparatus.pdf