This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Peggy Skerratt/Sandbox 1
From Proteopedia
| Line 4: | Line 4: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
==Overview== | ==Overview== | ||
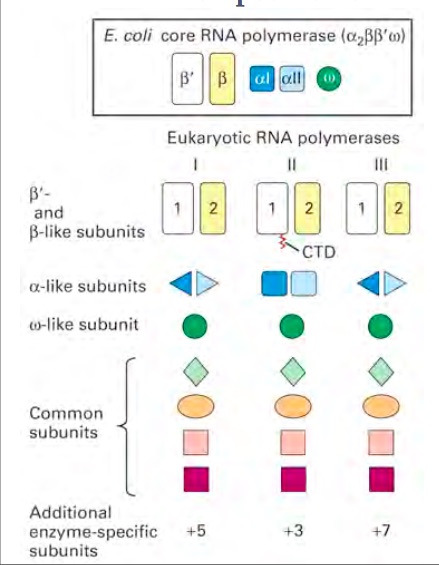
| - | RNA polymerase is an enzyme that binds to double stranded DNA and synthesizes RNA. Since the enzyme moves along the DNA strand to synthesise it, it is considered a molecular machine. RNA polymerase is similar to the DNA polymerase because it reads the DNA strand from the 3’ end to the 5’ end. It synthesizes the RNA from the 5’ end to the 3’ end. It only reads the template strand to synthesize the complementary strand. Unlike the DNA polymerase it does not need a primer to begin, it can initiate synthesis itself.<ref | + | RNA polymerase is an enzyme that binds to double stranded DNA and synthesizes RNA. Since the enzyme moves along the DNA strand to synthesise it, it is considered a molecular machine. RNA polymerase is similar to the DNA polymerase because it reads the DNA strand from the 3’ end to the 5’ end. It synthesizes the RNA from the 5’ end to the 3’ end. It only reads the template strand to synthesize the complementary strand. Unlike the DNA polymerase it does not need a primer to begin, it can initiate synthesis itself. <ref:1>This enzyme is found in both prokaryotes and eukaryotes. There are several different types of RNA polymerases. In Prokaryotes there is only one RNA polymerase, which synthesises all of the RNA. In eukaryotes there are three polymerases (RNA polymerase I,II, and III) that synthesize different types of RNA. Prokaryote and eukaryote polymerases have similar structures. Within the eukaryotic polymerases, the enzymes are structurally similar but have different subunits attached to the polymerase. The whole process of making RNA from DNA is called transcription. Transcription has four stages: assembly, initiation, elongation and termination, in which RNA polymerase is involved. The polymerase that is found to be a key polymerase is polymerase II. The exact mechanism for the different polymerases is still being studied. Since bacteria only have one polymerase that produce RNA, it is often studied to try to understand how this process works.The process of transcription is also regulated. It is controlled through specificity factors, repressors, and activators. If the transcription factors are not regulated, oncogenic activity can occur. Overall, RNA polymerase is still being studied and research is still being conducted to uncover all of the ins and outs of RNA polymerase’s role in transcription. |
| - | + | ||
| Line 18: | Line 17: | ||
==Mechanism== | ==Mechanism== | ||
| - | RNA polymerase is activated when it binds to specific gene sequences called promoters.6 There are several different kinds and much research is still being performed to discover the sequences necessary for binding. The promoter regions range from positions -70 to +30 from the transcription start site in E. coli. The subunit of RNA polymerase in E. coli directs its binding to the promoter. Locations -10 and -35 are major binding sites and have consensus sequences— (5’)TATAAT(3’) and (5’)TTGACA(3’) respectively.6 One promoter in common among Pol II is the TATA box and the Inr sequence YYANTYY or YYANAYY.6 | + | RNA polymerase is activated when it binds to specific gene sequences called promoters.6 There are several different kinds and much research is still being performed to discover the sequences necessary for binding. The promoter regions range from positions -70 to +30 from the transcription start site in E. coli. The subunit of RNA polymerase in E. coli directs its binding to the promoter. Locations -10 and -35 are major binding sites and have consensus sequences— (5’)TATAAT(3’) and (5’)TTGACA(3’) respectively.6 One promoter in common among Pol II is the TATA box and the Inr sequence YYANTYY or YYANAYY.6 |
| - | + | ||
| - | Regulatory factors on RNA polymerase II aid in RNA transcription initiation by attracting several parts of the transcription machinery to a central promoter.7 The transcription factor TBP (TATA binding protein) recognizes the TATA box and TFIIF binds to RNA Polymerase II to guide it away from non-promoter DNA sequences and towards TBP.6 TFIIB binds to Pol II, which now has TFIIF bound to it. TFIIA aids in Poll II binding to the promoter sequence. TFIIE and TFIIH also bind.6 Now that the polymerase and its factors are all bound to the DNA template, the RNA polymerase is positioned into its Preinitiation Complex (PIC).7 The PIC is not yet active for transcription at this point.7 TFIIH is a helicase6 and melts 11-15 base pairs near the transcription site--the hydrogen bonds between the base pairs of the two DNA strands are broken and the helix unwinds(7). | + | = Assembly = |
| - | Initiation | + | |
| + | Regulatory factors on RNA polymerase II aid in RNA transcription initiation by attracting several parts of the transcription machinery to a central promoter.7 The transcription factor TBP (TATA binding protein) recognizes the TATA box and TFIIF binds to RNA Polymerase II to guide it away from non-promoter DNA sequences and towards TBP.6 TFIIB binds to Pol II, which now has TFIIF bound to it. TFIIA aids in Poll II binding to the promoter sequence. TFIIE and TFIIH also bind.6 Now that the polymerase and its factors are all bound to the DNA template, the RNA polymerase is positioned into its Preinitiation Complex (PIC).7 The PIC is not yet active for transcription at this point.7 TFIIH is a helicase6 and melts 11-15 base pairs near the transcription site--the hydrogen bonds between the base pairs of the two DNA strands are broken and the helix unwinds(7). | ||
| + | Figure. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1189732/ (7) | ||
| + | |||
| + | = Initiation = | ||
| + | |||
In order to form the Open Complex, a conformational change occurs. A different subunit of TFIIH and protein kinase CDK9 phosphorylate Pol II at its C-terminal repeat domain (CTD) (6). Synthesis of RNA can now occur. Some organisms first complete abortive initiation, which is the formation of a few oligomeric mRNA strands(8). This is thought to occur due to little access to nucleotides substrates complementary to the promoter sequence(8) or instability of the RNAP II-DNA complex (9). | In order to form the Open Complex, a conformational change occurs. A different subunit of TFIIH and protein kinase CDK9 phosphorylate Pol II at its C-terminal repeat domain (CTD) (6). Synthesis of RNA can now occur. Some organisms first complete abortive initiation, which is the formation of a few oligomeric mRNA strands(8). This is thought to occur due to little access to nucleotides substrates complementary to the promoter sequence(8) or instability of the RNAP II-DNA complex (9). | ||
Once Pol II has synthesized an mRNA strand of at least 60 nucleotides long, TFIIE and TFIIH dissociate form the complex and Pol II leaves the promoter and begins elongation(6). Many of the transcription factors are left behind at the promoter to mark it as transcribed and to speed up the process for the next cycle--reinitiation of transcription. The complex left behind is called the Scaffold Complex.(7) | Once Pol II has synthesized an mRNA strand of at least 60 nucleotides long, TFIIE and TFIIH dissociate form the complex and Pol II leaves the promoter and begins elongation(6). Many of the transcription factors are left behind at the promoter to mark it as transcribed and to speed up the process for the next cycle--reinitiation of transcription. The complex left behind is called the Scaffold Complex.(7) | ||
| - | + | ||
| - | During Elongation, RNA polymerase covers 40 base pairs of DNA. Seventeen to 19 of those base pairs are unwound and create a transcription bubble, at which the new RNA is formed (8). Forming the transcription bubble requires energy from ATP and the presence of TFIIH and TFIIE (9). Nucleotides are then added, complementary to the DNA template strand, creating a DNA-RNA hybrid. RNAP II has two active sites for binding DNA--at the beginning and end of the polymerase (8). The polymerase alternates between being locked into place on the DNA strand, as nucleotides are added to the mRNA, and sliding along the template to the next transcription site.(8) The 3’ end on the newly synthesized mRNA binds to RNA polymerase at two points as well. | + | = Elongation = |
| - | The catalytic site of RNA polymerase II has 2 Mg(2+) ions coordinated with 3 aspartate residues in the polymerase structure. The Mg ions aid in the 3’ OH group on the growing RNA strand to attack the alpha-phosphate group of the incoming NTP and displacing the pyrophosphate. The presence of a specific nucleotide in the polymerase insertion site affects the geometry of the polymerase, which creates a pocket for the incoming nucleotide that is very specific to its shape. Therefore, base pairing rules and the shape of the active site influences the accuracy of elongation (6). | + | |
| + | During Elongation, RNA polymerase covers 40 base pairs of DNA. Seventeen to 19 of those base pairs are unwound and create a transcription bubble, at which the new RNA is formed (8). Forming the transcription bubble requires energy from ATP and the presence of TFIIH and TFIIE (9). Nucleotides are then added, complementary to the DNA template strand, creating a DNA-RNA hybrid. RNAP II has two active sites for binding DNA--at the beginning and end of the polymerase (8). The polymerase alternates between being locked into place on the DNA strand, as nucleotides are added to the mRNA, and sliding along the template to the next transcription site.(8) The 3’ end on the newly synthesized mRNA binds to RNA polymerase at two points as well. | ||
| + | The catalytic site of RNA polymerase II has 2 Mg(2+) ions coordinated with 3 aspartate residues in the polymerase structure. The Mg ions aid in the 3’ OH group on the growing RNA strand to attack the alpha-phosphate group of the incoming NTP and displacing the pyrophosphate. The presence of a specific nucleotide in the polymerase insertion site affects the geometry of the polymerase, which creates a pocket for the incoming nucleotide that is very specific to its shape. Therefore, base pairing rules and the shape of the active site influences the accuracy of elongation (6). | ||
Elongation factors are proteins bound to the phosphorylated CTD subunit of Pol II, which causes transcription to occur faster by limiting the transcription blocks(6). Some transcription blocks are resolved by the polymerase itself (pause), while others require the presence of certain elongation factors before transcription can continue (arrest)(9). When the 3’ OH of the RNA strand is not correctly aligned in the polymerase active site, a pause in transcription occurs (9). Transcriptional factors such as IIF, FCPI, CSB, and DSIF are involved in correcting for them. They act by increasing the rate of transcription (9). A transcriptional pause can turn into an arrest if the RNA polymerase stays at the block site long enough (9). Elongation factors allow the polymerase to overcome the arrest site, by facilitating the cleavage of the mRNA transcript, which realigns its position in the polymerase catalytic site (9). The elongation factor for eukaryotes is TFIIS, and for E.coli is GreA and GreB (9). | Elongation factors are proteins bound to the phosphorylated CTD subunit of Pol II, which causes transcription to occur faster by limiting the transcription blocks(6). Some transcription blocks are resolved by the polymerase itself (pause), while others require the presence of certain elongation factors before transcription can continue (arrest)(9). When the 3’ OH of the RNA strand is not correctly aligned in the polymerase active site, a pause in transcription occurs (9). Transcriptional factors such as IIF, FCPI, CSB, and DSIF are involved in correcting for them. They act by increasing the rate of transcription (9). A transcriptional pause can turn into an arrest if the RNA polymerase stays at the block site long enough (9). Elongation factors allow the polymerase to overcome the arrest site, by facilitating the cleavage of the mRNA transcript, which realigns its position in the polymerase catalytic site (9). The elongation factor for eukaryotes is TFIIS, and for E.coli is GreA and GreB (9). | ||
| - | + | ||
| - | Messenger RNA have a 3’ cap called a poly (A) tail composed of 80-250 adenosine residues (6). Termination of eukaryotic Pol II can occur 0.5 - 2 kb past the beginning of this tail (10). The mRNA is then cleaved at the addition site of the adenosine tail by a protein which is associated with the phosphorylated CTD subunit of Pol II (6). The cleavage site is marked by the (5’) AAUAAA (3’) sequence upstream and GU-rich region downstream (6). Polyadenylate polymerase adds more adenosine residues to the free 3’ end of the newly synthesized mRNA (6). | + | = Termination = |
| + | |||
| + | Messenger RNA have a 3’ cap called a poly (A) tail composed of 80-250 adenosine residues (6). Termination of eukaryotic Pol II can occur 0.5 - 2 kb past the beginning of this tail (10). The mRNA is then cleaved at the addition site of the adenosine tail by a protein which is associated with the phosphorylated CTD subunit of Pol II (6). The cleavage site is marked by the (5’) AAUAAA (3’) sequence upstream and GU-rich region downstream (6). Polyadenylate polymerase adds more adenosine residues to the free 3’ end of the newly synthesized mRNA (6). | ||
| + | == Headline text == | ||
| + | |||
| + | |||
== Regulation == | == Regulation == | ||
| - | Though regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site. | + | Though regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site[11]. |
== Connections with Cancer == | == Connections with Cancer == | ||
| - | RNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur. | + | RNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur[11]. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | < | + | www.youtube.com/watch?v=gqEwq4HnRmU. |
| + | Hurwitz, Jerard. “The Discovery of RNA Polymerase.” Journal of Biological Chemistry, 30 Dec. 2005, www.jbc.org/content/280/52/42477.'''<ref/>''' | ||
| + | Fredericks , Mike. “Prokaryotic vs Eukaryotic Transcription.” PROKARYOTIC VS EUKARYOTIC, 2006, www.chem.uwec.edu/webpapers2006/sites/fredermr/comp.html. | ||
| + | Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. Transcription and RNA polymerase. Available from: https://www.ncbi.nlm.nih.gov/books/NBK22085/ | ||
| + | Alber , T, et al. General Biochemistry and Molecular Biology . 22 Aug. 2008, mcb.berkeley.edu/courses/mcb110/ZHOU/lec.5-7.euk_trxn_apparatus.pdf | ||
Revision as of 23:22, 25 April 2018
[[Image:]]==RNA Polymerase==
| |||||||||||
References
www.youtube.com/watch?v=gqEwq4HnRmU.
Hurwitz, Jerard. “The Discovery of RNA Polymerase.” Journal of Biological Chemistry, 30 Dec. 2005, www.jbc.org/content/280/52/42477.Cite error: Invalid <ref> tag;
refs with no content must have a name
Fredericks , Mike. “Prokaryotic vs Eukaryotic Transcription.” PROKARYOTIC VS EUKARYOTIC, 2006, www.chem.uwec.edu/webpapers2006/sites/fredermr/comp.html.
Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. Transcription and RNA polymerase. Available from: https://www.ncbi.nlm.nih.gov/books/NBK22085/
Alber , T, et al. General Biochemistry and Molecular Biology . 22 Aug. 2008, mcb.berkeley.edu/courses/mcb110/ZHOU/lec.5-7.euk_trxn_apparatus.pdf