This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Khadar Abdi/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Threonyl-tRNA Synthetase/ligase== | ==Threonyl-tRNA Synthetase/ligase== | ||
| - | <StructureSection load=' | + | <StructureSection load='1NYQ' size='400' side='right' caption='Staphylococcus aureus threonyl-tRNA Synthetase bound to Threonyl-Sulfamoyl Adenosine' scene=''> |
| - | == Function/Mechanism == | + | == General Function/Mechanism == |
| - | '''Threonyl t-RNA Synthetase''' or '''Threonyl-tRNA ligase''' or '''TARS''' is class II '''Aminoacyl-tRNA synthetase''' enzymes. These enzymes primary function are to added the respective amino acid to the respective transfer Ribonucleic Acid (tRNA-AA) The main function of | + | '''Threonyl t-RNA Synthetase''' or '''Threonyl-tRNA ligase''' or '''TARS''' is class II '''Aminoacyl-tRNA synthetase''' enzymes. These enzymes primary function are to added the respective amino acid to the respective transfer Ribonucleic Acid (tRNA-AA) The main function of TARS is to add Threonine amino acid (Thr) to threonine specific tRNA (tRNA-thr) a necessity prep for the protein synthesis pathway. Below displays the overview of the Aminoacylation rxn. <ref>PMID:29305884</ref>[[Image:TARS_protein_rxn.jpeg |left|thumb|500px| '''Overall TARS protein rxn. Substrates includes Adenosine triphosphate (ATP), Threonine (Thr) and threonine specific transfer Ribonucleic Acid (tRNA-thr).''']] |
{{Clear}} | {{Clear}} | ||
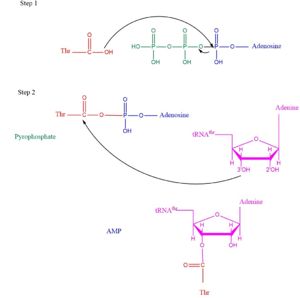
| - | TARS adds amino acid to tRNA by a two-step mechanism. First the enzyme binds to both <scene name='78/786634/Threonine_amino_acid_2/2'>Threonine</scene> and <scene name='78/786634/Atp/1'>ATP</scene> in the catalytic domain to perform an adenylation reaction in which pyrophosphate is released as a byproduct. This is then follow up by a transferring Thr from Adenosine monophosphate molecule to 3'OH site of tRNA-thr. <ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> | + | TARS adds amino acid to tRNA by a two-step mechanism. First the enzyme binds to both <scene name='78/786634/Threonine_amino_acid_2/2'>Threonine</scene> and <scene name='78/786634/Atp/1'>ATP</scene> in the catalytic domain to perform an adenylation reaction in which pyrophosphate is released as a byproduct. The image to the right displays the binding of adenylate product to the TARS enzyme (PDB entry [[1nyq]]). This is then follow up by a transferring Thr from Adenosine monophosphate molecule to 3'OH site of tRNA-thr. <ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> The image below demonstrates the arrow pushing occurring to generate threonine bound tRNA-thr. |
[[Image:TARSmechanism.jpg|left|thumb|300px|'''Arrow pushing of Aminoacylation rxn.<ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> ''']] | [[Image:TARSmechanism.jpg|left|thumb|300px|'''Arrow pushing of Aminoacylation rxn.<ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> ''']] | ||
| + | |||
| + | Lately the aaRS family was found to have more function than just aminoacylation. For instance, many aaRs molecules have been found to link with angiogenesis, blood vessel growth occuring within a cancer environment <ref>PMID:25535072</ref>. This is seen in human exogenous TARS in its ability to generate blood vessels within ovarian cancer environment <ref>PMID:27847344</ref>. Studies on the structure of TARS bound to BC194, derivative to the natural antibiotic Borrelidin, were investigated to understand how the angiogenic signaling from TARS occurs. | ||
== Structural highlights== | == Structural highlights== | ||
| - | == | + | == Evolutionary related proteins == |
== List to available structures == | == List to available structures == | ||
Revision as of 00:52, 29 April 2018
Threonyl-tRNA Synthetase/ligase
| |||||||||||