User:Khadar Abdi/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='1NYQ' size='400' side='right' caption='Staphylococcus aureus threonyl-tRNA Synthetase bound to Threonyl-Sulfamoyl Adenosine' scene=''> | <StructureSection load='1NYQ' size='400' side='right' caption='Staphylococcus aureus threonyl-tRNA Synthetase bound to Threonyl-Sulfamoyl Adenosine' scene=''> | ||
| - | == | + | == Introduction == |
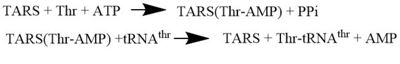
| - | '''Threonyl t-RNA Synthetase''' or '''Threonyl-tRNA ligase''' or '''TARS''' is class II '''Aminoacyl-tRNA synthetase''' enzymes. These enzymes primary function are to | + | '''Threonyl t-RNA Synthetase''' or '''Threonyl-tRNA ligase''' or '''TARS''' is a homodimer (150kDa in bacteria and 170kDa in human) and is classified as a class II '''Aminoacyl-tRNA synthetase''' enzymes. These ancient enzymes primary function are to add the respective amino acid to the respective transfer Ribonucleic Acid (tRNA-AA), a necessity prep for the protein synthesis pathway<ref>PMID:29305884</ref>. As the name implies, TARS function is to add Threonine amino acid (Thr) to threonine specific tRNA (tRNA-thr) in the presence of Adenosine triphosphate (ATP) and diavalent metal cation. Below displays the overview of TARS Aminoacylation rxn<ref>PMID:29305884</ref>.[[Image:TARS_protein_rxn.jpeg |center|thumb|400px| '''Overall TARS protein rxn. Substrates includes ATP, Thr and tRNA-thr.''']] |
{{Clear}} | {{Clear}} | ||
| - | + | ==Mechanism== | |
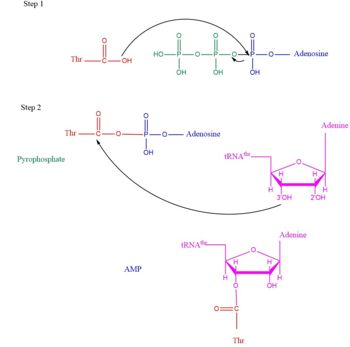
| - | [[Image:TARSmechanism.jpg| | + | [[Image:TARSmechanism.jpg|center|thumb|350px|'''Arrow pushing of Aminoacylation rxn.<ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> ''']] |
| - | Lately the aaRS family was found to have more function than just aminoacylation. For instance, many aaRs molecules have been found to link with angiogenesis, blood vessel growth occuring within a cancer environment <ref>PMID:25535072</ref>. This is seen in human exogenous TARS in its ability to generate blood vessels within ovarian cancer environment <ref>PMID: | + | TARS adds amino acid to tRNA by a two-step mechanism. First the enzyme binds to both <scene name='78/786634/Threonine_amino_acid_2/2'>Thr</scene> and <scene name='78/786634/Atp/1'>ATP</scene> in the catalytic domain to perform an adenylation reaction in which threonyl adenylate (Thr-AMP) is formed and pyrophosphate (PPi) is released as a byproduct. The image to the right displays the binding of adenylate product to the TARS enzyme (PDB entry [[1nyq]]). This is then follow up by a transferring Thr from Adenosine monophosphate (AMP) molecule to 3'OH site of tRNA-thr. <ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> The image to the above demonstrates the arrow pushing occurring to generate threonine bound tRNA-thr. |
| + | |||
| + | ==Disease Relevance== | ||
| + | |||
| + | Lately the aaRS family was found to have more function than just aminoacylation. For instance, many aaRs molecules have been found to link with angiogenesis, blood vessel growth occuring within a cancer environment<ref>PMID:25535072</ref>. This is seen in human exogenous TARS in its ability to generate blood vessels within ovarian cancer environment<ref>PMID:23425968</ref>. Studies on the structure of TARS bound to BC194, derivative to the natural antibiotic Borrelidin, were investigated to understand how the angiogenic signaling from TARS occurs<ref>PMID: 26271225</ref>. | ||
== Structural highlights== | == Structural highlights== | ||
| Line 17: | Line 21: | ||
== List to available structures == | == List to available structures == | ||
| + | |||
== References == | == References == | ||
Revision as of 01:31, 29 April 2018
Threonyl-tRNA Synthetase/ligase
| |||||||||||