Dioxygenase
From Proteopedia
(Difference between revisions)
| Line 56: | Line 56: | ||
**[[5umh]] – CTD – ''Burkholderia multivorans''<br /> | **[[5umh]] – CTD – ''Burkholderia multivorans''<br /> | ||
**[[5td3]] – CTD – ''Burkholderia vietnamiensis''<br /> | **[[5td3]] – CTD – ''Burkholderia vietnamiensis''<br /> | ||
| + | **[[5vxt]] – BaCTD + Fe+3 + catechol – ''Burkholderia ambifaria''<br /> | ||
*''CTD complex with aromatic compound'' | *''CTD complex with aromatic compound'' | ||
| Line 82: | Line 83: | ||
**[[4zxa]] – PsHQD + Cd+2 + benzonitrile <br /> | **[[4zxa]] – PsHQD + Cd+2 + benzonitrile <br /> | ||
**[[4zxc]] – PsHQD + Fe+3 <br /> | **[[4zxc]] – PsHQD + Fe+3 <br /> | ||
| + | **[[5m22]] – SpHQD + Fe+3 <br /> | ||
| + | **[[5m4o]] – SpHQD + Fe+3 + nitrophenol<br /> | ||
| + | **[[5m26]] – SpHQD + Fe+3 + hydroquinone derivative<br /> | ||
| + | **[[5m21]] – SpHQD + Fe+3 + hydroxybenzoate<br /> | ||
*Nitrobenzene dioxygenase (NBDO) | *Nitrobenzene dioxygenase (NBDO) | ||
| Line 110: | Line 115: | ||
**[[5f8p]], [[5dab]] – hFto + Fe+2 + oxoglutarate + inhibitor<br /> | **[[5f8p]], [[5dab]] – hFto + Fe+2 + oxoglutarate + inhibitor<br /> | ||
**[[4zs2]], [[4zs3]] – hFto + Mn+2 + oxoglutarate + fluorescin<br /> | **[[4zs2]], [[4zs3]] – hFto + Mn+2 + oxoglutarate + fluorescin<br /> | ||
| + | **[[6eoz]] – EnFto + Ni+2 + cyclopeptin – ''Emericela nidulans''<br /> | ||
| + | **[[5ybl]] – EnFto + Mn+2 + oxoglutarate <br /> | ||
| + | **[[5y7t]], [[5y7r]] – EnFto + Fe+3 + oxoglutarate + cyclopeptin<br /> | ||
| + | **[[5oa8]], [[5oa7]], [[5oa4]] – EnFto (mutant) + Ni+2 + oxoglutarate + cyclopeptin<br /> | ||
*''Alkb complex with DNA'' | *''Alkb complex with DNA'' | ||
| Line 282: | Line 291: | ||
**[[4xf3]] – rCysDO (mutant) + Fe+3 + inhibitor + cysteine<br /> | **[[4xf3]] – rCysDO (mutant) + Fe+3 + inhibitor + cysteine<br /> | ||
**[[2q4s]], [[2atf]] – mCysDO + Ni+2 <br /> | **[[2q4s]], [[2atf]] – mCysDO + Ni+2 <br /> | ||
| + | **[[6bgm]], [[6bgf]] – hCysDO + Fe+2 <br /> | ||
**[[2ic1]] – hCysDO + Fe+2 + cysteine<br /> | **[[2ic1]] – hCysDO + Fe+2 + cysteine<br /> | ||
| Line 315: | Line 325: | ||
**[[3swt]] – HAD + Fe+3 – ''Mycobacterium marinum''<br /> | **[[3swt]] – HAD + Fe+3 – ''Mycobacterium marinum''<br /> | ||
**[[5keu]] – BxHAD + Mg+2<br /> | **[[5keu]] – BxHAD + Mg+2<br /> | ||
| + | **[[5vn6]] – BaHAD + Mg+2<br /> | ||
*3-hydroxyanthranilate 3,4-dioxygenase (HAD) | *3-hydroxyanthranilate 3,4-dioxygenase (HAD) | ||
| Line 330: | Line 341: | ||
**[[3fe5]] – HAD + Fe+3 - bovine<br /> | **[[3fe5]] – HAD + Fe+3 - bovine<br /> | ||
**[[4wzc]] – HAD + Fe+2 + enedioate derivative – ''Ralstonia metallidurans''<br /> | **[[4wzc]] – HAD + Fe+2 + enedioate derivative – ''Ralstonia metallidurans''<br /> | ||
| + | **[[5v28]] – CmHAD + Fe+2 – ''Cupriavidus metallidurans''<br /> | ||
| + | **[[5v27]], [[5v26]] – CmHAD (mutant) + Fe+2 <br /> | ||
*Carbazole 1,9a-dioxygenase (CDO) | *Carbazole 1,9a-dioxygenase (CDO) | ||
| Line 355: | Line 368: | ||
**[[3rzz]] – SvPHPD (mutant) + Cd+2 <br /> | **[[3rzz]] – SvPHPD (mutant) + Cd+2 <br /> | ||
**[[3gbf]] – SvPHPD + Cd+2 + hydroxyethylphosphonate <br /> | **[[3gbf]] – SvPHPD + Cd+2 + hydroxyethylphosphonate <br /> | ||
| + | **[[6b9r]] – PHPD + Fe+3 + hydroxyethylphosphonate - ''Streptomyces albus''<br /> | ||
*Gallate dioxygenase (GD) | *Gallate dioxygenase (GD) | ||
| Line 380: | Line 394: | ||
**[[4qb5]] – GBD – ''Albidiferax ferrireducens''<br /> | **[[4qb5]] – GBD – ''Albidiferax ferrireducens''<br /> | ||
**[[4rt5]] – GBD – ''Planctomyces limnophilus''<br /> | **[[4rt5]] – GBD – ''Planctomyces limnophilus''<br /> | ||
| - | |||
**[[5uhj]], [[5ujp]] – GBD + Ni+2 – ''Streptomyces''<br /> | **[[5uhj]], [[5ujp]] – GBD + Ni+2 – ''Streptomyces''<br /> | ||
**[[5c4p]], [[5c68]], [[5cb9]] – TcGBD + Ni+2 – ''Thermomonospora curvata''<br /> | **[[5c4p]], [[5c68]], [[5cb9]] – TcGBD + Ni+2 – ''Thermomonospora curvata''<br /> | ||
| Line 404: | Line 417: | ||
* Indoleamine 2,3-dioxygenase (IAD) | * Indoleamine 2,3-dioxygenase (IAD) | ||
| - | **[[5ek2]], [[5ek3]], [[5ek4]], [[4pk5]] – hIAD + heme + imidazole derivative<br /> | + | **[[6azu]] – hIAD + heme <br /> |
| + | **[[5ek2]], [[5ek3]], [[5ek4]], [[4pk5]], [[5xe1]], [[5wn8]] – hIAD + heme + imidazole derivative<br /> | ||
**[[4u72]], [[4u74]] – hIAD (mutant) + heme + imidazole derivative<br /> | **[[4u72]], [[4u74]] – hIAD (mutant) + heme + imidazole derivative<br /> | ||
| + | **[[6f0a]] – hIAD + heme + imidazole derivative + Ala<br /> | ||
| + | **[[6azw]], [[6azv]] – hIAD + inhibitor<br /> | ||
| + | **[[5etw]] – hIAD + heme + inhibitor<br /> | ||
| + | **[[5wmu]] – hIAD + heme + CN + Trp<br /> | ||
| + | **[[5wmw]] – hIAD (mutant) + heme + CN + Trp<br /> | ||
| + | **[[5wmv]] – hIAD + heme + CN + indole derivative + Trp<br /> | ||
| + | **[[5wmx]] – hIAD (mutant) + heme + CN + indole derivative + Trp<br /> | ||
| + | **[[5whr]] – hIAD + heme + pyrrolydine derivative <br /> | ||
* Thymine dioxygenase (TD) | * Thymine dioxygenase (TD) | ||
| Line 413: | Line 435: | ||
**[[5c3q]] – NcTD + oxoglutarate + Ni+2 + thymine<br /> | **[[5c3q]] – NcTD + oxoglutarate + Ni+2 + thymine<br /> | ||
**[[5c3s]] – NcTD + oxoglutarate + Ni+2 + pyrimidine derivative <br /> | **[[5c3s]] – NcTD + oxoglutarate + Ni+2 + pyrimidine derivative <br /> | ||
| - | **[[ | + | **[[5c3r]] – NcTD + oxoglutarate + Ni+2 + pyrimidine derivative + uracil derivative<br /> |
* 2,4’-dihydroxyacetophenone dioxygenase (APD) | * 2,4’-dihydroxyacetophenone dioxygenase (APD) | ||
Revision as of 07:44, 1 May 2018
| |||||||||||
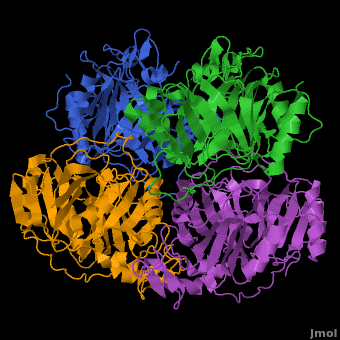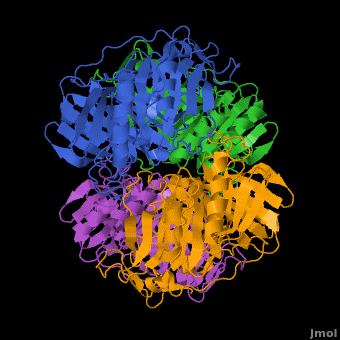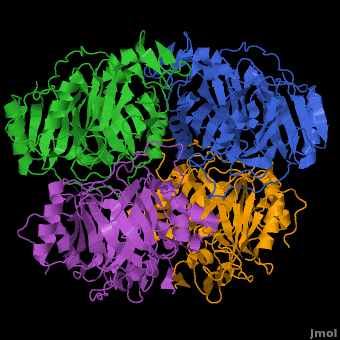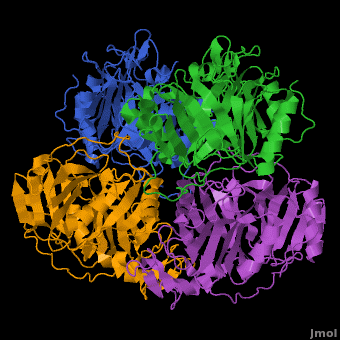
3D structures of Protocatechuate 3,4-dioxygenase
Updated on 01-May-2018
- ↑ Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, Tian Q, Aifantis I, Wei L, Dong C. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015 Apr 21;42(4):613-26. doi: 10.1016/j.immuni.2015.03.005. Epub 2015 , Apr 7. PMID:25862091 doi:http://dx.doi.org/10.1016/j.immuni.2015.03.005
- ↑ Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L Jr. A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase. J Biol Inorg Chem. 2010 Dec 14. PMID:21153851 doi:10.1007/s00775-010-0732-0
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky