User:Khadar Abdi/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Introduction == | == Introduction == | ||
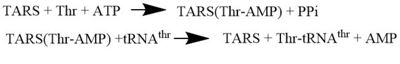
'''Threonyl t-RNA Synthetase''' or '''Threonyl-tRNA ligase''' or '''TARS''' is a homodimer (150kDa in bacteria and 170kDa in human) and is classified as a class II '''Aminoacyl-tRNA synthetase''' enzymes. These ancient enzymes primary function are to add the respective amino acid to the respective transfer Ribonucleic Acid (tRNA-AA), a necessity prep for the protein synthesis pathway<ref>PMID:29305884</ref>. As the name implies, TARS function is to add Threonine amino acid (Thr) to threonine specific tRNA (tRNA-thr) in the presence of Adenosine triphosphate (ATP) and diavalent metal cation. Below displays the overview of TARS Aminoacylation rxn<ref>PMID:29305884</ref>.[[Image:TARS_protein_rxn.jpeg |center|thumb|400px| '''Overall TARS protein rxn. Substrates includes ATP, Thr and tRNA-thr.''']] | '''Threonyl t-RNA Synthetase''' or '''Threonyl-tRNA ligase''' or '''TARS''' is a homodimer (150kDa in bacteria and 170kDa in human) and is classified as a class II '''Aminoacyl-tRNA synthetase''' enzymes. These ancient enzymes primary function are to add the respective amino acid to the respective transfer Ribonucleic Acid (tRNA-AA), a necessity prep for the protein synthesis pathway<ref>PMID:29305884</ref>. As the name implies, TARS function is to add Threonine amino acid (Thr) to threonine specific tRNA (tRNA-thr) in the presence of Adenosine triphosphate (ATP) and diavalent metal cation. Below displays the overview of TARS Aminoacylation rxn<ref>PMID:29305884</ref>.[[Image:TARS_protein_rxn.jpeg |center|thumb|400px| '''Overall TARS protein rxn. Substrates includes ATP, Thr and tRNA-thr.''']] | ||
| + | |||
| + | Although there are multiple forms of TARS in different cells, this protopedia page mainly focuses on the structure of ''Staphylococcus aureus'' (''S. aureus'') and ''Escherichia coli'' (''E. coli'') cytoplasmic TARS as their is more crystal structures that identify their domains within the protein. | ||
| Line 11: | Line 13: | ||
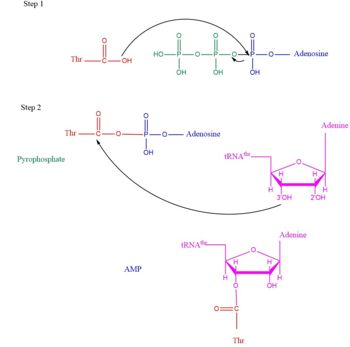
TARS adds amino acid to tRNA by a two-step mechanism. First the enzyme binds to both <scene name='78/786634/Threonine_amino_acid_2/2'>Thr</scene> and <scene name='78/786634/Atp/1'>ATP</scene> in the catalytic domain to perform an adenylation reaction in which threonyl adenylate (Thr-AMP) is formed and pyrophosphate (PPi) is released as a byproduct. The image to the right displays the binding of adenylate product to the TARS enzyme (PDB entry [[1nyq]]). This is then follow up by a transferring Thr from Adenosine monophosphate (AMP) molecule to 3'OH site of tRNA-thr. <ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> The image to the above demonstrates the arrow pushing occurring to generate threonine bound tRNA-thr. | TARS adds amino acid to tRNA by a two-step mechanism. First the enzyme binds to both <scene name='78/786634/Threonine_amino_acid_2/2'>Thr</scene> and <scene name='78/786634/Atp/1'>ATP</scene> in the catalytic domain to perform an adenylation reaction in which threonyl adenylate (Thr-AMP) is formed and pyrophosphate (PPi) is released as a byproduct. The image to the right displays the binding of adenylate product to the TARS enzyme (PDB entry [[1nyq]]). This is then follow up by a transferring Thr from Adenosine monophosphate (AMP) molecule to 3'OH site of tRNA-thr. <ref>Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). ''Lehninger principles of biochemistry.'' New York: Worth Publishers.</ref> The image to the above demonstrates the arrow pushing occurring to generate threonine bound tRNA-thr. | ||
| - | |||
| - | ==Disease Relevance== | ||
| - | |||
| - | Lately the aaRS family was found to have more function than just aminoacylation. For instance, many aaRs molecules have been found to link with angiogenesis, blood vessel growth occuring within a cancer environment<ref>PMID:25535072</ref>. This is seen in human exogenous TARS in its ability to generate blood vessels within ovarian cancer environment<ref>PMID:23425968</ref>. Studies on the structure of TARS bound to BC194, derivative to the natural antibiotic Borrelidin, were investigated to understand how the angiogenic signaling from TARS occurs<ref>PMID: 26271225</ref>. | ||
== Structural highlights== | == Structural highlights== | ||
| - | As mentioned earlier, TARS is homodimer protein found in two forms of a cell, mitochondrial and cytoplasmic. Most of the structures seen will primarly be cytoplasmic. The protein is mainly classified as a alpha beta protein in both bacteria and eukaryotic cells<ref>PMID:23362938</ref>. Each chain is composed of 4 domain classified by SCOP: a N1 domain, a N2 domain, a catalytic domain and an anti-codon domain<ref>PMID:12875846</ref>.<Structure load='Image of ThrS Staph colored label by domains' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | + | |
| + | As mentioned earlier, TARS is homodimer protein found in two forms of a cell, mitochondrial and cytoplasmic. Most of the structures seen will primarly be bacterial cytoplasmic. The protein is mainly classified as a alpha beta protein in both bacteria and eukaryotic cells<ref>PMID:23362938</ref>. Each chain is composed of 4 domain classified by SCOP: a N1 domain, a N2 domain, a catalytic domain and an anti-codon domain<ref>PMID:12875846</ref>.<Structure load='Image of ThrS Staph colored label by domains' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | ||
===N-terminus (Residue 1-241)=== | ===N-terminus (Residue 1-241)=== | ||
| Line 24: | Line 23: | ||
This is the opposite of the N2-domain, or sometimes called TARS additional domain. This is ironic as this addition is very important for proofreading TARS activity<ref>PMID:15525511</ref>. TARS N2-domain is described as 2-layer alpha-beta sandwich comprised of mostly alpha helices. Interesting enough, the superfamily of this motif is found to be TARS and Alanyl-tRNA synthetase common domain. This domain function similar by discriminating Threonine and Alanine from Serine amino acid (Ser) as serine is similar structure to both amino acids. The editing domain functions first by moving Ser bound tRNA-Thr from the catalytic domain to the editing site by breaking the bonds nucleotide A73, C74, and C75 from the catalytic site allowing the acceptor arm of tRNA-Thr to flip to the editing domain of S12-S13 site. Research on editing hydrolysis propose that residue Tyr 103 plays an important role of guiding acceptor arm towards the editing domain. The Ser bound to tRNA is hydrolzed by a water molecule, interacting with His73, acting as a nucleophile to the alpha carbon of serine, follow by protonation by a 2nd water molecule interacting with carbonyl of Met181 and amide side chain Lys156. The fidility mechanism of AA-tRNA binding is much similar to alanine tRNA-ligase found in the C-terminus as they share 40% similarity in residues. The subtle difference of the isoforms mitochondrial TARS from cytoplasmic TARS appears in editing as the mitochondrial TARS doesn't have N2-domain and requires interaction to hydrolyze serine <ref>PMID:22773845</ref>. | This is the opposite of the N2-domain, or sometimes called TARS additional domain. This is ironic as this addition is very important for proofreading TARS activity<ref>PMID:15525511</ref>. TARS N2-domain is described as 2-layer alpha-beta sandwich comprised of mostly alpha helices. Interesting enough, the superfamily of this motif is found to be TARS and Alanyl-tRNA synthetase common domain. This domain function similar by discriminating Threonine and Alanine from Serine amino acid (Ser) as serine is similar structure to both amino acids. The editing domain functions first by moving Ser bound tRNA-Thr from the catalytic domain to the editing site by breaking the bonds nucleotide A73, C74, and C75 from the catalytic site allowing the acceptor arm of tRNA-Thr to flip to the editing domain of S12-S13 site. Research on editing hydrolysis propose that residue Tyr 103 plays an important role of guiding acceptor arm towards the editing domain. The Ser bound to tRNA is hydrolzed by a water molecule, interacting with His73, acting as a nucleophile to the alpha carbon of serine, follow by protonation by a 2nd water molecule interacting with carbonyl of Met181 and amide side chain Lys156. The fidility mechanism of AA-tRNA binding is much similar to alanine tRNA-ligase found in the C-terminus as they share 40% similarity in residues. The subtle difference of the isoforms mitochondrial TARS from cytoplasmic TARS appears in editing as the mitochondrial TARS doesn't have N2-domain and requires interaction to hydrolyze serine <ref>PMID:22773845</ref>. | ||
| - | ===Catalytic Domain (Residue 243-=== | + | ===Catalytic Domain (Residue 243-532)=== |
| - | ===Anti-Codon Domain=== | + | As the name implies, the catalytic domain is the main area of protein activity. This domain is linked with N2-Domain of TARS by a alpha helix, or a linker helix. The domain is architecturally described as a 2-layer alpha beta sandwhich. The importance of the domain is that it has 3 sites of binding: motif 1: ordering loop; motif 2 loop, Motif 3: threonine loop, and motif 4: ATP binding. The main important residues within in the caatalytic site include: Tyr ,Arg , Arg , His309 |
| + | |||
| + | |||
| + | What is interesting about the motif 1 is that it is highly dependent on the presence of zinc divalent ion (Zn2+). The Zn2+ plays a huge salt bridge role for connecting Thr and TARS itself <ref>PMID:10319817</ref>. Zn2+ binds to TARS 2 histidine and 1 cysteine residue (for staph it was Cys336, His387, and His571) and the Thr amino acid's hydroxyl side chain and N-terminus. This prevents similar molecules lacking hydroxyl groups such as valine. Asides Zn2+, Mg2+ is also necessary for | ||
| + | |||
| + | ===Anti-Codon Domain(Residue 533-645)=== | ||
| + | |||
| + | This domain resides at the C-terminus end of the TARS protein. | ||
== Evolutionary related proteins == | == Evolutionary related proteins == | ||
| + | |||
| + | ==Disease Relevance== | ||
| + | |||
| + | Lately the aaRS family was found to have more function than just aminoacylation. For instance, many aaRs molecules have been found to link with angiogenesis, blood vessel growth occuring within a cancer environment<ref>PMID:25535072</ref>. This is seen in human exogenous TARS in its ability to generate blood vessels within ovarian cancer environment<ref>PMID:23425968</ref>. Studies on the structure of TARS bound to BC194, derivative to the natural antibiotic Borrelidin, were investigated to understand how the angiogenic signaling from TARS occurs<ref>PMID: 26271225</ref>. | ||
== List to available structures == | == List to available structures == | ||
| + | ===Bacterial=== | ||
| + | E coli: [[1QF6]] | ||
| + | S. Aureus: [[1NYQ]] | ||
| + | Yeast: [[3UGT]] | ||
| + | ===Eukaryotic=== | ||
| + | Human: [[4hwt]](w/ L-threonimide inhibitor); [[4ttv]] (w/ BC194 inhibitor) | ||
== References == | == References == | ||
Revision as of 19:01, 1 May 2018
Threonyl-tRNA Synthetase/ligase
| |||||||||||