We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Christian Fjeld/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
== General Description == | == General Description == | ||
<StructureSection load='4aq7' frame='true' size='340' side='right' caption='LARS (E coli) ternary complex with tRNAleu and leucyl adenylate analogue' scene='78/786656/Lars/2'> | <StructureSection load='4aq7' frame='true' size='340' side='right' caption='LARS (E coli) ternary complex with tRNAleu and leucyl adenylate analogue' scene='78/786656/Lars/2'> | ||
| - | <scene name='78/786656/Total/1'>Leucyl tRNA synthetase (LARS)</scene> is a 97 kDa, class IA aminoacyl-tRNA synthetase (ARS) that catalyzes the ligation of leucine with tRNA<sup>leu</sup> in an ATP dependent mechanism. LARS is a cytoplasmic enzyme that is found as part of the multisynthetase complex in eukaryotes<ref>doi: 10.1007/978-3-319-46503-6_18</ref>. The multi-synthetase complex contains glutamylprolyl-tRNA synthetase (EPRS), isoleucyl (IARS), leucyl (LARS), glutaminyl (GARS), methionyl (MARS), lysyl (KARS), arginyl (RARS), and aspartyl (DARS) tRNA synthetases as well as p18, p38 and p43<ref>doi: 10.1016/S0006-291X(03)00485-6</ref>. | + | <scene name='78/786656/Total/1'>Leucyl tRNA synthetase (LARS)</scene> is a 97 kDa, class IA aminoacyl-tRNA synthetase (ARS) that catalyzes the ligation of leucine with tRNA<sup>leu</sup> in an ATP dependent mechanism. ARS are essential to all life as they charge amino acids onto cognate tRNAs in preparation for protein translation. This is step is a potential source of error in interpreting the genetic code as mischarged tRNAs will not be recognized during protein synthesis and could lead to nonfunctional proteins. As such, ARS have a high specificity for both tRNA and amino acids and contain an editing domain capable of hydrolyzing mischarged tRNA. |
| + | |||
| + | LARS is a cytoplasmic enzyme that is found as part of the multisynthetase complex in eukaryotes<ref>doi: 10.1007/978-3-319-46503-6_18</ref>. The multi-synthetase complex contains glutamylprolyl-tRNA synthetase (EPRS), isoleucyl (IARS), leucyl (LARS), glutaminyl (GARS), methionyl (MARS), lysyl (KARS), arginyl (RARS), and aspartyl (DARS) tRNA synthetases as well as p18, p38 and p43<ref>doi: 10.1016/S0006-291X(03)00485-6</ref>. | ||
Multisynthetase complexes have also been seen in some archaea such as ''Thermococcus kodakarensis'' although the composition of the complex is not the same as eukaryotes<ref>doi: 10.1016/j.febslet.2012.05.039</ref>. | Multisynthetase complexes have also been seen in some archaea such as ''Thermococcus kodakarensis'' although the composition of the complex is not the same as eukaryotes<ref>doi: 10.1016/j.febslet.2012.05.039</ref>. | ||
LARS has been shown to be involved with the mTOR pathways as a sensor of leucine levels within the cell<ref>doi: 10.1016/j.cell.2012.02.044</ref>. | LARS has been shown to be involved with the mTOR pathways as a sensor of leucine levels within the cell<ref>doi: 10.1016/j.cell.2012.02.044</ref>. | ||
| + | |||
| + | === Disease === | ||
Mutations in LARS2, the mitochondrial version of the enzyme, have been linked to Perrault syndrome characterized by premature ovarian failure in females and progressive hearing loss in both sexes<ref>doi: 10.1016/j.ajhg.2013.03.007</ref> | Mutations in LARS2, the mitochondrial version of the enzyme, have been linked to Perrault syndrome characterized by premature ovarian failure in females and progressive hearing loss in both sexes<ref>doi: 10.1016/j.ajhg.2013.03.007</ref> | ||
| Line 10: | Line 14: | ||
=== Catalytic Domain === | === Catalytic Domain === | ||
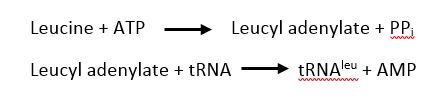
| - | The <scene name='78/786656/Catalytic/1'>catalytic domain</scene> is responsible for the two step process of charging leucine on to tRNA<sup>leu</sup>. First, ATP and leucine are bound and AMP is transfered to the backbone carboxylic acid of leucine with the release of a pyrophosphate. Second, tRNA<sup>leu</sup> is bound with the leucyl adenylate and leucine is transfered to either the 2 | + | |
| + | The <scene name='78/786656/Catalytic/1'>catalytic domain</scene> is responsible for the two step process of charging leucine on to tRNA<sup>leu</sup>. First, ATP and leucine are bound and AMP is transfered to the backbone carboxylic acid of leucine with the release of a pyrophosphate. Second, tRNA<sup>leu</sup> is bound with the leucyl adenylate and leucine is transfered to either the 2' OH of the 3' terminal adenine with the release of AMP<ref>doi: 10.1038/nsmb.2317</ref>. | ||
[[Image:Scheme.JPG]] | [[Image:Scheme.JPG]] | ||
| + | |||
| + | LARS, like all class I synthetases, is characterized by a Rossmann-fold catalytic domain with a central parallel β-sheet with α-helices on both faces. The active site catalyzes both the formation of the leucyl adenylate intermediate and the subsequent charging of leucine onto the terminal acceptor arm of the tRNA. | ||
<ref>doi: 10.1093/emboj/19.10.2351</ref> | <ref>doi: 10.1093/emboj/19.10.2351</ref> | ||
| - | <ref>doi: 10.1038/nsmb.2317</ref> | ||
=== Editing Domain === | === Editing Domain === | ||
| + | |||
The <scene name='78/786656/Editing/1'>editing domain</scene> of LARS is responsible for editing mischarged tRNA and ensuring translational fidelity. | The <scene name='78/786656/Editing/1'>editing domain</scene> of LARS is responsible for editing mischarged tRNA and ensuring translational fidelity. | ||
<ref>doi: 10.1016/j.jmb.2009.04.073</ref> | <ref>doi: 10.1016/j.jmb.2009.04.073</ref> | ||
| Line 23: | Line 30: | ||
=== Anticodon Binding Domain === | === Anticodon Binding Domain === | ||
| + | |||
The <scene name='78/786656/Anticodon/1'>anticodon binding domain</scene> is essential for the fidelity of ARSs. However, there are numerous anticodons that correspond to leucine including AAU, GUA, and GUG | The <scene name='78/786656/Anticodon/1'>anticodon binding domain</scene> is essential for the fidelity of ARSs. However, there are numerous anticodons that correspond to leucine including AAU, GUA, and GUG | ||
=== Zinc Binding Domain === | === Zinc Binding Domain === | ||
| + | |||
The <scene name='78/786656/Zn1/1'>zinc binding domain</scene> | The <scene name='78/786656/Zn1/1'>zinc binding domain</scene> | ||
=== Leucine Specificity Domain === | === Leucine Specificity Domain === | ||
| + | |||
The <scene name='78/786656/Ls/1'>leucine specificity domain</scene> | The <scene name='78/786656/Ls/1'>leucine specificity domain</scene> | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
| + | |||
A <scene name='78/786656/Conservation/2'>map of the conserved residues</scene> | A <scene name='78/786656/Conservation/2'>map of the conserved residues</scene> | ||
{{Template:ColorKey_ConSurf}} | {{Template:ColorKey_ConSurf}} | ||
| - | == Disease == | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== 3D Structure of LARS == | == 3D Structure of LARS == | ||
| + | |||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
{{#tree:id=OrganizedByTopic|openlevels=0| | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
Revision as of 02:17, 2 May 2018
General Description
| |||||||||||
3D Structure of LARS
Updated on 02-May-2018
References
- ↑ Mirande M. The Aminoacyl-tRNA Synthetase Complex. Subcell Biochem. 2017;83:505-522. doi: 10.1007/978-3-319-46503-6_18. PMID:28271488 doi:http://dx.doi.org/10.1007/978-3-319-46503-6_18
- ↑ Han JM, Kim JY, Kim S. Molecular network and functional implications of macromolecular tRNA synthetase complex. Biochem Biophys Res Commun. 2003 Apr 18;303(4):985-93. doi: , 10.1016/s0006-291x(03)00485-6. PMID:12684031 doi:http://dx.doi.org/10.1016/s0006-291x(03)00485-6
- ↑ Raina M, Elgamal S, Santangelo TJ, Ibba M. Association of a multi-synthetase complex with translating ribosomes in the archaeon Thermococcus kodakarensis. FEBS Lett. 2012 Jul 30;586(16):2232-8. doi: 10.1016/j.febslet.2012.05.039. Epub, 2012 Jun 7. PMID:22683511 doi:http://dx.doi.org/10.1016/j.febslet.2012.05.039
- ↑ Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012 Apr 13;149(2):410-24. doi: 10.1016/j.cell.2012.02.044. Epub 2012 Mar, 15. PMID:22424946 doi:http://dx.doi.org/10.1016/j.cell.2012.02.044
- ↑ Pierce SB, Gersak K, Michaelson-Cohen R, Walsh T, Lee MK, Malach D, Klevit RE, King MC, Levy-Lahad E. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am J Hum Genet. 2013 Apr 4;92(4):614-20. doi: 10.1016/j.ajhg.2013.03.007. Epub, 2013 Mar 28. PMID:23541342 doi:http://dx.doi.org/10.1016/j.ajhg.2013.03.007
- ↑ Palencia A, Crepin T, Vu MT, Lincecum TL Jr, Martinis SA, Cusack S. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat Struct Mol Biol. 2012 Jun 10. doi: 10.1038/nsmb.2317. PMID:22683997 doi:10.1038/nsmb.2317
- ↑ Cusack S, Yaremchuk A, Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000 May 15;19(10):2351-61. PMID:10811626 doi:10.1093/emboj/19.10.2351
- ↑ Seiradake E, Mao W, Hernandez V, Baker SJ, Plattner JJ, Alley MR, Cusack S. Crystal structures of the human and fungal cytosolic Leucyl-tRNA synthetase editing domains: A structural basis for the rational design of antifungal benzoxaboroles. J Mol Biol. 2009 Jul 10;390(2):196-207. Epub 2009 May 6. PMID:19426743 doi:10.1016/j.jmb.2009.04.073
- ↑ Liu Y, Liao J, Zhu B, Wang ED, Ding J. Crystal structures of the editing domain of Escherichia coli leucyl-tRNA synthetase and its complexes with Met and Ile reveal a lock-and-key mechanism for amino acid discrimination. Biochem J. 2006 Mar 1;394(Pt 2):399-407. PMID:16277600 doi:10.1042/BJ20051249