We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Aminomutase
From Proteopedia
(Difference between revisions)
| Line 32: | Line 32: | ||
== Structural highlights == | == Structural highlights == | ||
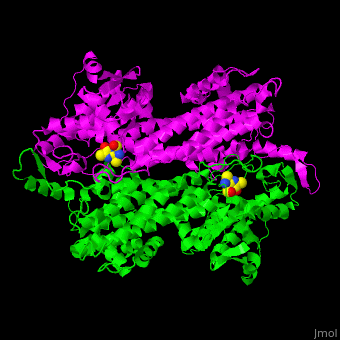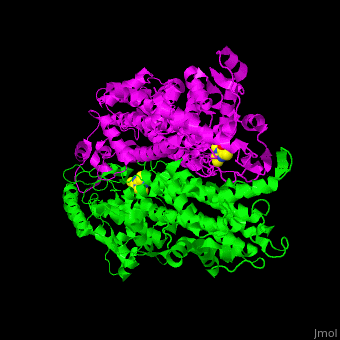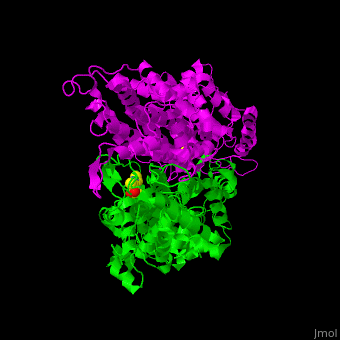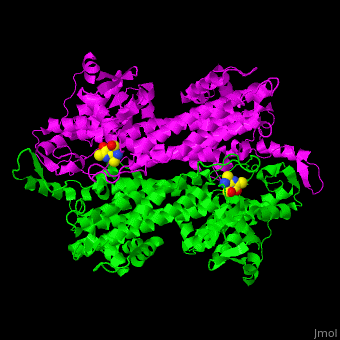
| - | TAM active site residues facilitate the deprotonation/reprotonation reaction. <scene name='71/715468/Cv/ | + | TAM active site residues facilitate the deprotonation/reprotonation reaction. <scene name='71/715468/Cv/3'>The active site</scene> includes residues from both monomers in Tyrosine aminomutase (PDB code [[2ohy]]).<ref name="Ad">PMID:17516659</ref> |
</StructureSection> | </StructureSection> | ||
Revision as of 12:07, 2 May 2018
| |||||||||||
3D structures of aminomutase
Updated on 02-May-2018
References
- ↑ 1.0 1.1 Christianson CV, Montavon TJ, Van Lanen SG, Shen B, Bruner SD. The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry. 2007 Jun 19;46(24):7205-14. Epub 2007 May 22. PMID:17516659 doi:10.1021/bi7003685