This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Patrick Wiencek/AHNAK
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== '''Introduction''' == | == '''Introduction''' == | ||
| - | <scene name='78/786654/4ftgjustahnak/2'>AHNAK</scene> is a protein characterized by its large size (700 kDa) and its unique tripartite structure <ref name="a1">PMID:25172424</ref><ref name="a2">PMID:8408266</ref>. Originally identified in 1989 as a desmosomal plaque protein purified from bovine muzzle epidermis called desmoyokin, AHNAK is now recognized as a scaffolding protein that has implicated in a wide range of diverse biological processes <ref name="a1" /><ref name="a2" />. This includes processes from Ca2+ channel regulation and cell adhesion to cell cycle arrest <ref name="a1" /><sup>[3-5]</sup>. Despite the diversity of processes that AHNAK is involved in, they’re unified in the formation of multi-protein complexes, AHNAK likely serving as a scaffolding protein for other proteins in the complex < | + | <scene name='78/786654/4ftgjustahnak/2'>AHNAK</scene> is a protein characterized by its large size (700 kDa) and its unique tripartite structure <ref name="a1">PMID:25172424</ref><ref name="a2">PMID:8408266</ref>. Originally identified in 1989 as a desmosomal plaque protein purified from bovine muzzle epidermis called desmoyokin, AHNAK is now recognized as a scaffolding protein that has implicated in a wide range of diverse biological processes <ref name="a1" /><ref name="a2" />. This includes processes from Ca2+ channel regulation and cell adhesion to cell cycle arrest <ref name="a1" /><sup>[3-5]</sup>. Despite the diversity of processes that AHNAK is involved in, they’re unified in the formation of multi-protein complexes, AHNAK likely serving as a scaffolding protein for other proteins in the complex <ref name="a1" />. |
| - | In alignment with AHNAK’s many functions, AHNAK has several identified subcellular localizations depending on cell type, intercellular contacts, and the phosphorylation state of the protein <sup>[ | + | In alignment with AHNAK’s many functions, AHNAK has several identified subcellular localizations depending on cell type, intercellular contacts, and the phosphorylation state of the protein <ref name="a1" /><sup>[6]</sup>. These include the plasma membrane, the cytoplasm and the nucleus <ref name="a2" /><sup>[7-9]</sup>. There is also evidence supporting AHNAK’s export to the extracellular space <sup>[9]</sup>. |
== '''Structure''' == | == '''Structure''' == | ||
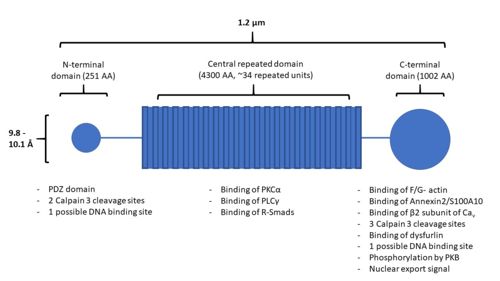
| - | AHNAK has a unique structure made up of three main domains: the N-terminal domain, the central repeated domain, and the C-terminal domain (Figure 1). These domains are 251, 4300, and 1002 amino acids in length, respectively <sup>[ | + | AHNAK has a unique structure made up of three main domains: the N-terminal domain, the central repeated domain, and the C-terminal domain (Figure 1). These domains are 251, 4300, and 1002 amino acids in length, respectively <ref name="a1" /><sup>[8]</sup>. |
| - | [[Image:AHNAKFigure1.1.jpg|500px|right|thumb|Figure 1. A structural representation of AHNAK, including sites of protein interaction. Modified from | + | [[Image:AHNAKFigure1.1.jpg|500px|right|thumb|Figure 1. A structural representation of AHNAK, including sites of protein interaction. Modified from <ref name="a1" />]] |
=== The Central Repeated Domain === | === The Central Repeated Domain === | ||
| Line 51: | Line 51: | ||
The MH2 domain of Smad2 will bind to the central repetitive domain of AHNAK from residues 4105-4633 <sup>[21]</sup>. | The MH2 domain of Smad2 will bind to the central repetitive domain of AHNAK from residues 4105-4633 <sup>[21]</sup>. | ||
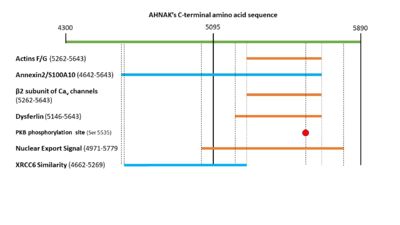
| - | It is also of not that AHNAK does not have a calcium binding domain, despite it responding to calcium signaling. Calcium sensing might be facilitated by AHNAK’s interaction with annexin 2, which is calcium sensitive <sup>[ | + | It is also of not that AHNAK does not have a calcium binding domain, despite it responding to calcium signaling. Calcium sensing might be facilitated by AHNAK’s interaction with annexin 2, which is calcium sensitive <ref name="a1" /><sup>[22,23]</sup>. |
== '''Function''' == | == '''Function''' == | ||
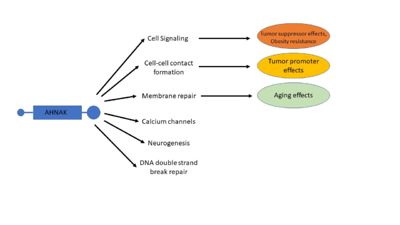
| - | AHNAK has a diverse list of biological processes that is has been implicated in, including cell signaling and cell contacts, regulation of calcium channels, membrane repair, and interaction with DNA ligase (Figure 4) < | + | AHNAK has a diverse list of biological processes that is has been implicated in, including cell signaling and cell contacts, regulation of calcium channels, membrane repair, and interaction with DNA ligase (Figure 4) <ref name="a1" />. AHNAK has been implicated in each of these biological processes, each with a description of its role: |
| - | [[Image:AHNAKFigure2.1.jpg|400px|right|thumb|Figure 4. A visual representation of many of AHNAK's functions and how those functions relate to disease. Modified from | + | [[Image:AHNAKFigure2.1.jpg|400px|right|thumb|Figure 4. A visual representation of many of AHNAK's functions and how those functions relate to disease. Modified from <ref name="a1" />]] |
=== Cell signaling === | === Cell signaling === | ||
| Line 69: | Line 69: | ||
=== Cell-Cell contact formation === | === Cell-Cell contact formation === | ||
| - | In addition to phosphorylation by PKB, AHNAK localization in epithelial cells depends on cell confluency, where sub-confluent cells displayed a nuclear localization while confluent cells displayed a cytoplasmic or plasma membrane localization <sup>[6]</sup>. When AHNAK re-localizes to the plasma membrane it complexes with actin and heterotetrameric annexin2/S100A10 <sup>[7]</sup>. A structural analysis of this complex reveals that both annexin2 and S100A10 are necessary for the complex to form <sup>[29]</sup>. A possible mechanism for calcium dependent cell-cell contact formation is that PKB phosphorylation of AHNAK will cause its translocation to the plasma membrane where it complexes with actin and annexin2/S100A10 <sup>[ | + | In addition to phosphorylation by PKB, AHNAK localization in epithelial cells depends on cell confluency, where sub-confluent cells displayed a nuclear localization while confluent cells displayed a cytoplasmic or plasma membrane localization <sup>[6]</sup>. When AHNAK re-localizes to the plasma membrane it complexes with actin and heterotetrameric annexin2/S100A10 <sup>[7]</sup>. A structural analysis of this complex reveals that both annexin2 and S100A10 are necessary for the complex to form <sup>[29]</sup>. A possible mechanism for calcium dependent cell-cell contact formation is that PKB phosphorylation of AHNAK will cause its translocation to the plasma membrane where it complexes with actin and annexin2/S100A10 <ref name="a1" /><sup>[6,7]</sup>. |
=== Calcium channels === | === Calcium channels === | ||
| - | AHNAK can bind the β2 subunit of L-type voltage gated calcium (Cav ) channels in cardiomyocytes <sup>[13]</sup>. AHNAK seems to have different effects on calcium channels and from calcium across the cited studies. This may be due to different calcium channel isoforms, or different cell types (and thus different responses to calcium) < | + | AHNAK can bind the β2 subunit of L-type voltage gated calcium (Cav ) channels in cardiomyocytes <sup>[13]</sup>. AHNAK seems to have different effects on calcium channels and from calcium across the cited studies. This may be due to different calcium channel isoforms, or different cell types (and thus different responses to calcium) <ref name="a1" />. One hypothesis of AHNAK function with the β2 subunit is that following β-adrenergic stimulation and phosphorylation of AHNAK by PKA, AHNAK will release the β2 subunit of the Cav channel and allow normal calcium influx <sup>[31]</sup>. AHNAK was also implicated in calcium influx in CD4+ T cells and cytotoxic CD8+ effector T-cells <sup>[32,33]</sup>. Here, AHNAK null mice showed decreased calcium influx, leading experts to hypothesize that the underlying mechanism involved AHNAK assisting the β2-subunit in membrane localization <sup>[34]</sup>. |
=== Membrane repair === | === Membrane repair === | ||
| Line 84: | Line 84: | ||
== '''AHNAK in Disease''' == | == '''AHNAK in Disease''' == | ||
| - | Despite initial mouse models that showed no phenotypic defects in AHNAK-null mice, AHNAK has been related to several different diseases <sup>[10,37]</sup>. These include but are not limited to: cancer, obesity, and aging <sup>[ | + | Despite initial mouse models that showed no phenotypic defects in AHNAK-null mice, AHNAK has been related to several different diseases <sup>[10,37]</sup>. These include but are not limited to: cancer, obesity, and aging <ref name="a1" /><sup>[38-40]</sup>. |
=== Cancer === | === Cancer === | ||
| Line 90: | Line 90: | ||
AHNAK’s roles in cancer and tumor metastasis have recently become a large part of the research being done with AHNAK. Due to AHNAK’s implications in many different biological processes, AHNAK seems to promote cancer in some contexts <sup>[30,41]</sup>, and serve as a tumor suppressor in others <sup>[3,4,21]</sup>. Due to its functionality in cytoskeletal stabilization and interaction with actin filaments, AHNAK was found to be essential in actin-rich pseudopod protrusion across several different metastatic human tumor cell lines <sup>[30]</sup>. AHNAK knockdown caused these cells to retract their pseudopods and reverse the epithelial to mesenchymal transition that is necessary for cancer metastasis <sup>[30]</sup>. Similarly, significantly higher levels of AHNAK expression were detected in mesotheliomal cell lines, and migration and invasion were both decreased following AHNAK knockdown <sup>[41]</sup>. | AHNAK’s roles in cancer and tumor metastasis have recently become a large part of the research being done with AHNAK. Due to AHNAK’s implications in many different biological processes, AHNAK seems to promote cancer in some contexts <sup>[30,41]</sup>, and serve as a tumor suppressor in others <sup>[3,4,21]</sup>. Due to its functionality in cytoskeletal stabilization and interaction with actin filaments, AHNAK was found to be essential in actin-rich pseudopod protrusion across several different metastatic human tumor cell lines <sup>[30]</sup>. AHNAK knockdown caused these cells to retract their pseudopods and reverse the epithelial to mesenchymal transition that is necessary for cancer metastasis <sup>[30]</sup>. Similarly, significantly higher levels of AHNAK expression were detected in mesotheliomal cell lines, and migration and invasion were both decreased following AHNAK knockdown <sup>[41]</sup>. | ||
| - | AHNAK can also act as a tumor suppressor because of its role in the TFGβ/Smad pathway <sup>[21]</sup>. Overexpression of AHNAK in mouse fibroblast cell resulted in increased cell-cycle arrest. Analysis of AHNAK mRNA levels in glioma demonstrated that AHNAK was down-regulated in some cell lines, and was a statistically significant prognostic factor for poor survival of glioma patients <sup>[4]</sup>. Similar results were shown in a study of AHNAK in triple-negative breast cancer, also associating AHNAK with the AMK/MAPK signaling pathway and the Wnt/β-catenin pathway <sup>[3]</sup>. These differing effects of AHNAK in cancer may involve its regulation via TGFβ, which has both tumor suppressor and tumor promotor roles <sup>[ | + | AHNAK can also act as a tumor suppressor because of its role in the TFGβ/Smad pathway <sup>[21]</sup>. Overexpression of AHNAK in mouse fibroblast cell resulted in increased cell-cycle arrest. Analysis of AHNAK mRNA levels in glioma demonstrated that AHNAK was down-regulated in some cell lines, and was a statistically significant prognostic factor for poor survival of glioma patients <sup>[4]</sup>. Similar results were shown in a study of AHNAK in triple-negative breast cancer, also associating AHNAK with the AMK/MAPK signaling pathway and the Wnt/β-catenin pathway <sup>[3]</sup>. These differing effects of AHNAK in cancer may involve its regulation via TGFβ, which has both tumor suppressor and tumor promotor roles <ref name="a1" /><sup>[42]</sup>. |
=== Obesity === | === Obesity === | ||
| Line 121: | Line 121: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | |||
| - | 1. Davis, T. A., Loos, B. & Engelbrecht, A.-M. AHNAK: the giant jack of all trades. Cell. Signal. 26, 2683–2693 (2014). | ||
| - | |||
| - | 2. Hashimoto, T. et al. Desmoyokin, a 680 kDa keratinocyte plasma membrane-associated protein, is homologous to the protein encoded by human gene AHNAK. J. Cell Sci. 105 ( Pt 2), 275–286 (1993). | ||
| - | |||
| - | 3. Chen, B. et al. AHNAK suppresses tumour proliferation and invasion by targeting multiple pathways in triple-negative breast cancer. J. Exp. Clin. Cancer Res. CR 36, 65 (2017). | ||
| - | |||
| - | 4. Zhao, Z. et al. AHNAK as a Prognosis Factor Suppresses the Tumor Progression in Glioma. J. Cancer 8, 2924–2932 (2017). | ||
| - | |||
| - | 5. Davis, T. et al. Doxorubicin resistance in breast cancer: A novel role for the human protein AHNAK. Biochem. Pharmacol. 148, 174–183 (2018). | ||
| - | |||
| - | 6. Sussman, J., Stokoe, D., Ossina, N. & Shtivelman, E. Protein kinase B phosphorylates AHNAK and regulates its subcellular localization. J. Cell Biol. 154, 1019–1030 (2001). | ||
| - | |||
| - | 7. Benaud, C. et al. AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. J. Cell Biol. 164, 133–144 (2004). | ||
| - | |||
| - | 8. Shtivelman, E., Cohen, F. E. & Bishop, J. M. A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc. Natl. Acad. Sci. U. S. A. 89, 5472–5476 (1992). | ||
| - | |||
| - | 9. Cell atlas - AHNAK - The Human Protein Atlas. Available at: http://www.proteinatlas.org/ENSG00000124942-AHNAK/cell. (Accessed: 30th April 2018) | ||
| - | |||
| - | 10. Komuro, A. et al. The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc. Natl. Acad. Sci. 101, 4053–4058 (2004). | ||
| - | |||
| - | 11. Lee, H.-J. & Zheng, J. J. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signal. 8, 8 (2010). | ||
| - | |||
| - | 12. de Morrée, A. et al. Self-regulated alternative splicing at the AHNAK locus. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 26, 93–103 (2012). | ||
| - | |||
| - | 13. Hohaus, A. et al. The carboxyl-terminal region of ahnak provides a link between cardiac L-type Ca2+ channels and the actin-based cytoskeleton. FASEB J. 16, 1205–1216 (2002). | ||
| - | |||
| - | 14. Huang, Y. et al. Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle. Hum. Mol. Genet. 17, 1855–1866 (2008). | ||
| - | |||
| - | 15. Huang, Y. et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 21, 732–742 (2006). | ||
| - | |||
| - | 16. Stiff, T., Shtivelman, E., Jeggo, P. & Kysela, B. AHNAK interacts with the DNA ligase IV-XRCC4 complex and stimulates DNA ligase IV-mediated double-stranded ligation. DNA Repair 3, 245–256 (2004). | ||
| - | |||
| - | 17. EMBOSS Needle < Pairwise Sequence Alignment < EMBL-EBI. Available at: https://www.ebi.ac.uk/Tools/psa/emboss_needle/. (Accessed: 2nd May 2018) | ||
| - | |||
| - | 18. AHNAK - Neuroblast differentiation-associated protein AHNAK - Homo sapiens (Human) - AHNAK gene & protein. Available at: https://www.uniprot.org/uniprot/Q09666#ptm_processing. (Accessed: 1st May 2018) | ||
| - | |||
| - | 19. Lee, I. H. et al. Ahnak Protein Activates Protein Kinase C (PKC) through Dissociation of the PKC-Protein Phosphatase 2A Complex. J. Biol. Chem. 283, 6312–6320 (2008). | ||
| - | |||
| - | 20. Sekiya, F., Bae, Y. S., Jhon, D. Y., Hwang, S. C. & Rhee, S. G. AHNAK, a Protein That Binds and Activates Phospholipase C-γ1 in the Presence of Arachidonic Acid. J. Biol. Chem. 274, 13900–13907 (1999). | ||
| - | |||
| - | 21. Lee, I. H. et al. Ahnak functions as a tumor suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene 33, 4675–4684 (2014). | ||
| - | |||
| - | 22. Grieve, A. G., Moss, S. E. & Hayes, M. J. Annexin A2 at the Interface of Actin and Membrane Dynamics: A Focus on Its Roles in Endocytosis and Cell Polarization. International Journal of Cell Biology (2012). Available at: https://www.hindawi.com/journals/ijcb/2012/852430/. (Accessed: 2nd May 2018) | ||
| - | |||
| - | 23. Rezvanpour, A., Santamaria-Kisiel, L. & Shaw, G. S. The S100A10-Annexin A2 Complex Provides a Novel Asymmetric Platform for Membrane Repair. J. Biol. Chem. 286, 40174–40183 (2011). | ||
| - | |||
| - | 24. Chang, F. et al. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia 17, 1263–1293 (2003). | ||
| - | |||
| - | 25. Boxberg, Y. V. et al. Spinal cord injury-induced up-regulation of AHNAK, expressed in cells delineating cystic cavities, and associated with neoangiogenesis. Eur. J. Neurosci. 24, 1031–1041 (2006). | ||
| - | |||
| - | 26. Salim, C., Boxberg, Y. V., Alterio, J., Féréol, S. & Nothias, F. The giant protein AHNAK involved in morphogenesis and laminin substrate adhesion of myelinating Schwann cells. Glia 57, 535–549 (2009). | ||
| - | |||
| - | 27. Gentil, B. J. et al. Specific AHNAK expression in brain endothelial cells with barrier properties. J. Cell. Physiol. 203, 362–371 (2005). | ||
| - | |||
| - | 28. Shin, J. H. et al. Increased Cell Proliferations and Neurogenesis in the Hippocampal Dentate Gyrus of Ahnak Deficient Mice. Neurochem. Res. 40, 1457–1462 (2015). | ||
| - | |||
| - | 29. Dempsey, B. R. et al. Structure of an Asymmetric Ternary Protein Complex Provides Insight for Membrane Interaction. Structure 20, 1737–1745 (2012). | ||
| - | |||
| - | 30. Shankar, J. et al. Pseudopodial Actin Dynamics Control Epithelial-Mesenchymal Transition in Metastatic Cancer Cells. Cancer Res. 70, 3780–3790 (2010). | ||
| - | |||
| - | 31. Alvarez, J. et al. Calcium Current in Rat Cardiomyocytes Is Modulated by the Carboxyl-terminal Ahnak Domain. J. Biol. Chem. 279, 12456–12461 (2004). | ||
| - | |||
| - | 32. Matza, D. et al. A Scaffold Protein, AHNAK1, Is Required for Calcium Signaling during T Cell Activation. Immunity 28, 64–74 (2008). | ||
| - | |||
| - | 33. Matza, D. et al. Requirement for AHNAK1-mediated calcium signaling during T lymphocyte cytolysis. Proc. Natl. Acad. Sci. 106, 9785–9790 (2009). | ||
| - | |||
| - | 34. Chien, A. J. et al. Roles of a Membrane-localized βSubunit in the Formation and Targeting of Functional L-type Ca2+ Channels. J. Biol. Chem. 270, 30036–30044 (1995). | ||
| - | |||
| - | 35. Borgonovo, B. et al. Regulated exocytosis: a novel, widely expressed system. Nat. Cell Biol. 4, 955–963 (2002). | ||
| - | |||
| - | 36. Lennon, N. J. et al. Dysferlin Interacts with Annexins A1 and A2 and Mediates Sarcolemmal Wound-healing. J. Biol. Chem. 278, 50466–50473 (2003). | ||
| - | |||
| - | 37. Kouno, M. et al. Ahnak/Desmoyokin Is Dispensable for Proliferation, Differentiation, and Maintenance of Integrity in Mouse Epidermis. J. Invest. Dermatol. 123, 700–707 (2004). | ||
| - | |||
| - | 38. Su, J. et al. A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet. Muscle 5, 35 (2015). | ||
| - | |||
| - | 39. de Magalhães, J. P., Curado, J. & Church, G. M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinforma. Oxf. Engl. 25, 875–881 (2009). | ||
| - | |||
| - | 40. Parikh, H. et al. Molecular correlates for maximal oxygen uptake and type 1 fibers. Am. J. Physiol.-Endocrinol. Metab. 294, E1152–E1159 (2008). | ||
| - | |||
| - | 41. Sudo, H. et al. AHNAK is highly expressed and plays a key role in cell migration and invasion in mesothelioma. Int. J. Oncol. 44, 530–538 (2014). | ||
| - | |||
| - | 42. Heldin, C.-H., Landström, M. & Moustakas, A. Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial–mesenchymal transition. Curr. Opin. Cell Biol. 21, 166–176 (2009). | ||
| - | |||
| - | 43. Kim, I. Y. et al. 1H NMR-based metabolomic study on resistance to diet-induced obesity in AHNAK knock-out mice. Biochem. Biophys. Res. Commun. 403, 428–434 (2010). | ||
| - | |||
| - | 44. Shin, J. H. et al. Obesity Resistance and Enhanced Insulin Sensitivity in Ahnak -/- Mice Fed a High Fat Diet Are Related to Impaired Adipogenesis and Increased Energy Expenditure. PLoS ONE 10, (2015). | ||
| - | |||
| - | 45. AceView: Gene:AHNAK, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView. Available at: https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&c=Gene&l=AHNAK. (Accessed: 30th April 2018) | ||
| - | |||
| - | 46. Gillespie, C. S., Sherman, D. L., Blair, G. E. & Brophy, P. J. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron 12, 497–508 (1994). | ||
| - | |||
| - | 47. Dytrych, L., Sherman, D. L., Gillespie, C. S. & Brophy, P. J. Two PDZ Domain Proteins Encoded by the Murine Periaxin Gene Are the Result of Alternative Intron Retention and Are Differentially Targeted in Schwann Cells. J. Biol. Chem. 273, 5794–5800 (1998). | ||
| - | |||
| - | 48. Han, H. & Kursula, P. Periaxin and AHNAK nucleoprotein 2 form intertwined homodimers through domain swapping. J. Biol. Chem. 289, 14121–14131 (2014). | ||
| - | |||
| - | 49. Ozorowski, G., Milton, S. & Luecke, H. Structure of a C-terminal AHNAK peptide in a 1:2:2 complex with S100A10 and an acetylated N-terminal peptide of annexin A2. Acta Crystallogr. D Biol. Crystallogr. 69, 92–104 (2013). | ||
| - | |||
| - | 50. Oh, Y.-S. et al. SMARCA3, a chromatin-remodeling factor, is required for p11-dependent antidepressant action. Cell 152, 831–843 (2013). | ||
<references/> | <references/> | ||
Revision as of 20:05, 4 May 2018
AHNAK
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Davis TA, Loos B, Engelbrecht AM. AHNAK: the giant jack of all trades. Cell Signal. 2014 Dec;26(12):2683-93. doi: 10.1016/j.cellsig.2014.08.017. Epub, 2014 Aug 27. PMID:25172424 doi:http://dx.doi.org/10.1016/j.cellsig.2014.08.017
- ↑ 2.0 2.1 2.2 Hashimoto T, Amagai M, Parry DA, Dixon TW, Tsukita S, Tsukita S, Miki K, Sakai K, Inokuchi Y, Kudoh J, et al.. Desmoyokin, a 680 kDa keratinocyte plasma membrane-associated protein, is homologous to the protein encoded by human gene AHNAK. J Cell Sci. 1993 Jun;105 ( Pt 2):275-86. PMID:8408266