User:Patrick Wiencek/AHNAK
From Proteopedia
(Difference between revisions)
| Line 40: | Line 40: | ||
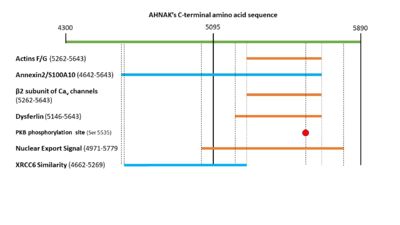
The N-terminal region of dysfurlin will interact with the C-terminal domain of AHNAK from residues 5146 – 5643 <ref name="a15">Huang, Y. et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 21, 732–742 (2006).</ref>. | The N-terminal region of dysfurlin will interact with the C-terminal domain of AHNAK from residues 5146 – 5643 <ref name="a15">Huang, Y. et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 21, 732–742 (2006).</ref>. | ||
*'''DNA''' | *'''DNA''' | ||
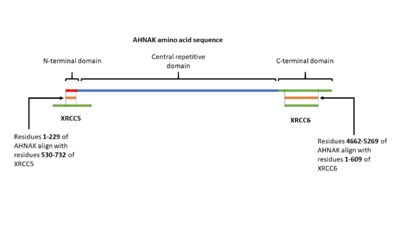
| - | AHNAK has been shown having weak DNA binding affinity similar to the Ku protein <ref name="a16">DOI:10.1016/j.dnarep.2003.11.001</ref>. Sequence alignment of AHNAK with Ku70 (Uniprot P13010)and Ku80 (Uniprot P12956) indicated areas of similarity from residues 1-200 and 4661-5260 respectively (Figure 3) <ref name=" | + | AHNAK has been shown having weak DNA binding affinity similar to the Ku protein <ref name="a16">DOI:10.1016/j.dnarep.2003.11.001</ref>. Sequence alignment of AHNAK with Ku70 (Uniprot P13010)and Ku80 (Uniprot P12956) indicated areas of similarity from residues 1-200 and 4661-5260 respectively (Figure 3) <ref name="a17">EMBOSS Needle < Pairwise Sequence Alignment < EMBL-EBI. Available at: https://www.ebi.ac.uk/Tools/psa/emboss_needle/. (Accessed: 2nd May 2018)</ref>. These sites may be AHNAK’s prospective DNA binding sites. |
[[Image:AHNAKFigureKu.2.jpg|400px|right|thumb|Figure 3. A visual representation of AHNAK's amino acid sequence and its sites of similarity with the proteins XRCC5 and XRCC6.]] | [[Image:AHNAKFigureKu.2.jpg|400px|right|thumb|Figure 3. A visual representation of AHNAK's amino acid sequence and its sites of similarity with the proteins XRCC5 and XRCC6.]] | ||
*'''Protein Kinase B (PKB)''' | *'''Protein Kinase B (PKB)''' | ||
| - | PKB will phosphorylate serine 5535 in AHNAK’s C-terminal domain <ref name="a6" />. This will activate AHNAK’s nuclear export signal, allowing it to move out of the nucleus. AHNAK’s nuclear export signal is made up of 5 different motifs in the C-terminal domain: (4971-4979), (5019-5027), (5034-5039), (5706-5716), and (5772-5779) <ref name=" | + | PKB will phosphorylate serine 5535 in AHNAK’s C-terminal domain <ref name="a6" />. This will activate AHNAK’s nuclear export signal, allowing it to move out of the nucleus. AHNAK’s nuclear export signal is made up of 5 different motifs in the C-terminal domain: (4971-4979), (5019-5027), (5034-5039), (5706-5716), and (5772-5779) <ref name="a18">AHNAK - Neuroblast differentiation-associated protein AHNAK - Homo sapiens (Human) - AHNAK gene & protein. Available at: https://www.uniprot.org/uniprot/Q09666#ptm_processing. (Accessed: 1st May 2018)</ref>. |
*'''Protein Kinase C α (PKCα)''' | *'''Protein Kinase C α (PKCα)''' | ||
PKCα will bind to and is activated by AHNAK <sup>[19]</sup>. This interaction occurs in AHNAK’s central repeated domain (3859-4412). | PKCα will bind to and is activated by AHNAK <sup>[19]</sup>. This interaction occurs in AHNAK’s central repeated domain (3859-4412). | ||
Revision as of 20:31, 4 May 2018
AHNAK
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Davis TA, Loos B, Engelbrecht AM. AHNAK: the giant jack of all trades. Cell Signal. 2014 Dec;26(12):2683-93. doi: 10.1016/j.cellsig.2014.08.017. Epub, 2014 Aug 27. PMID:25172424 doi:http://dx.doi.org/10.1016/j.cellsig.2014.08.017
- ↑ 2.0 2.1 2.2 Hashimoto T, Amagai M, Parry DA, Dixon TW, Tsukita S, Tsukita S, Miki K, Sakai K, Inokuchi Y, Kudoh J, et al.. Desmoyokin, a 680 kDa keratinocyte plasma membrane-associated protein, is homologous to the protein encoded by human gene AHNAK. J Cell Sci. 1993 Jun;105 ( Pt 2):275-86. PMID:8408266
- ↑ Chen B, Wang J, Dai D, Zhou Q, Guo X, Tian Z, Huang X, Yang L, Tang H, Xie X. AHNAK suppresses tumour proliferation and invasion by targeting multiple pathways in triple-negative breast cancer. J Exp Clin Cancer Res. 2017 May 12;36(1):65. doi: 10.1186/s13046-017-0522-4. PMID:28494797 doi:http://dx.doi.org/10.1186/s13046-017-0522-4
- ↑ Zhao Z, Xiao S, Yuan X, Yuan J, Zhang C, Li H, Su J, Wang X, Liu Q. AHNAK as a Prognosis Factor Suppresses the Tumor Progression in Glioma. J Cancer. 2017 Aug 25;8(15):2924-2932. doi: 10.7150/jca.20277. eCollection 2017. PMID:28928883 doi:http://dx.doi.org/10.7150/jca.20277
- ↑ Davis T, van Niekerk G, Peres J, Prince S, Loos B, Engelbrecht AM. Doxorubicin resistance in breast cancer: A novel role for the human protein AHNAK. Biochem Pharmacol. 2018 Feb;148:174-183. doi: 10.1016/j.bcp.2018.01.012. Epub, 2018 Jan 5. PMID:29309757 doi:http://dx.doi.org/10.1016/j.bcp.2018.01.012
- ↑ 6.0 6.1 Sussman J, Stokoe D, Ossina N, Shtivelman E. Protein kinase B phosphorylates AHNAK and regulates its subcellular localization. J Cell Biol. 2001 Sep 3;154(5):1019-30. doi: 10.1083/jcb.200105121. PMID:11535620 doi:http://dx.doi.org/10.1083/jcb.200105121
- ↑ 7.0 7.1 Benaud C, Gentil BJ, Assard N, Court M, Garin J, Delphin C, Baudier J. AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. J Cell Biol. 2004 Jan 5;164(1):133-44. doi: 10.1083/jcb.200307098. Epub 2003 Dec , 29. PMID:14699089 doi:http://dx.doi.org/10.1083/jcb.200307098
- ↑ 8.0 8.1 8.2 Shtivelman E, Cohen FE, Bishop JM. A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5472-6. PMID:1608957
- ↑ 9.0 9.1 Cell atlas - AHNAK - The Human Protein Atlas. Available at: http://www.proteinatlas.org/ENSG00000124942-AHNAK/cell. (Accessed: 30th April 2018)
- ↑ Komuro A, Masuda Y, Kobayashi K, Babbitt R, Gunel M, Flavell RA, Marchesi VT. The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc Natl Acad Sci U S A. 2004 Mar 23;101(12):4053-8. doi:, 10.1073/pnas.0308619101. Epub 2004 Mar 8. PMID:15007166 doi:http://dx.doi.org/10.1073/pnas.0308619101
- ↑ Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010 May 28;8:8. doi: 10.1186/1478-811X-8-8. PMID:20509869 doi:http://dx.doi.org/10.1186/1478-811X-8-8
- ↑ 12.0 12.1 de Morree A, Droog M, Grand Moursel L, Bisschop IJ, Impagliazzo A, Frants RR, Klooster R, van der Maarel SM. Self-regulated alternative splicing at the AHNAK locus. FASEB J. 2012 Jan;26(1):93-103. doi: 10.1096/fj.11-187971. Epub 2011 Sep 22. PMID:21940993 doi:http://dx.doi.org/10.1096/fj.11-187971
- ↑ Hohaus A, Person V, Behlke J, Schaper J, Morano I, Haase H. The carboxyl-terminal region of ahnak provides a link between cardiac L-type Ca2+ channels and the actin-based cytoskeleton. FASEB J. 2002 Aug;16(10):1205-16. doi: 10.1096/fj.01-0855com. PMID:12153988 doi:http://dx.doi.org/10.1096/fj.01-0855com
- ↑ Huang, Y. et al. Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle. Hum. Mol. Genet. 17, 1855–1866 (2008).
- ↑ Huang, Y. et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 21, 732–742 (2006).
- ↑ Stiff T, Shtivelman E, Jeggo P, Kysela B. AHNAK interacts with the DNA ligase IV-XRCC4 complex and stimulates DNA ligase IV-mediated double-stranded ligation. DNA Repair (Amst). 2004 Mar 4;3(3):245-56. doi: 10.1016/j.dnarep.2003.11.001. PMID:15177040 doi:http://dx.doi.org/10.1016/j.dnarep.2003.11.001
- ↑ EMBOSS Needle < Pairwise Sequence Alignment < EMBL-EBI. Available at: https://www.ebi.ac.uk/Tools/psa/emboss_needle/. (Accessed: 2nd May 2018)
- ↑ AHNAK - Neuroblast differentiation-associated protein AHNAK - Homo sapiens (Human) - AHNAK gene & protein. Available at: https://www.uniprot.org/uniprot/Q09666#ptm_processing. (Accessed: 1st May 2018)