2hue
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 2hue |SIZE=350|CAPTION= <scene name='initialview01'>2hue</scene>, resolution 1.7Å | |PDB= 2hue |SIZE=350|CAPTION= <scene name='initialview01'>2hue</scene>, resolution 1.7Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= ASF1 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 Saccharomyces cerevisiae]), H3l ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=8355 Xenopus laevis]) | |GENE= ASF1 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 Saccharomyces cerevisiae]), H3l ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=8355 Xenopus laevis]) | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY=[[1kx5|1KX5]], [[1roc|1ROC]] | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2hue FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2hue OCA], [http://www.ebi.ac.uk/pdbsum/2hue PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=2hue RCSB]</span> | ||
}} | }} | ||
| Line 26: | Line 29: | ||
[[Category: English, C M]] | [[Category: English, C M]] | ||
[[Category: Tyler, J K.]] | [[Category: Tyler, J K.]] | ||
| - | [[Category: GOL]] | ||
| - | [[Category: SO4]] | ||
| - | [[Category: ZN]] | ||
[[Category: elongated beta sandwhich]] | [[Category: elongated beta sandwhich]] | ||
[[Category: mini beta sheet]] | [[Category: mini beta sheet]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 03:35:32 2008'' |
Revision as of 00:35, 31 March 2008
| |||||||
| , resolution 1.7Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||
| Gene: | ASF1 (Saccharomyces cerevisiae), H3l (Xenopus laevis) | ||||||
| Related: | 1KX5, 1ROC
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
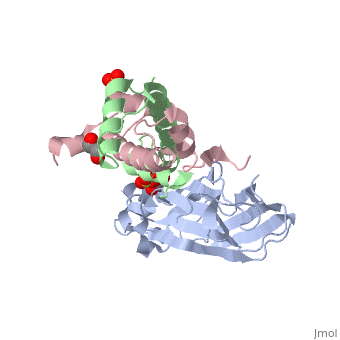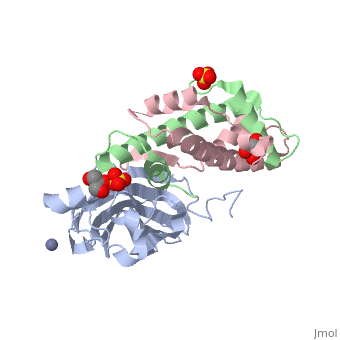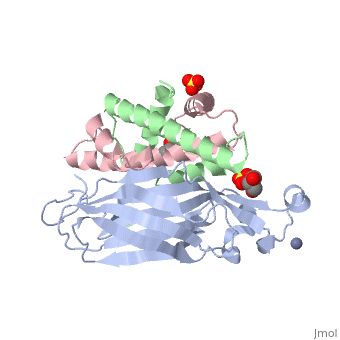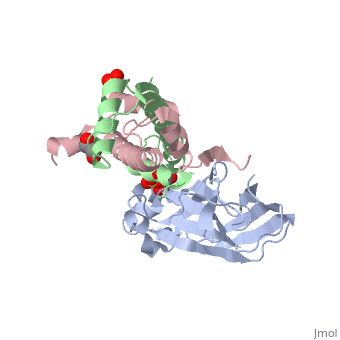
Structure of the H3-H4 chaperone Asf1 bound to histones H3 and H4
Overview
Anti-silencing function 1 (Asf1) is a highly conserved chaperone of histones H3/H4 that assembles or disassembles chromatin during transcription, replication, and repair. The structure of the globular domain of Asf1 bound to H3/H4 determined by X-ray crystallography to a resolution of 1.7 Angstroms shows how Asf1 binds the H3/H4 heterodimer, enveloping the C terminus of histone H3 and physically blocking formation of the H3/H4 heterotetramer. Unexpectedly, the C terminus of histone H4 that forms a mini-beta sheet with histone H2A in the nucleosome undergoes a major conformational change upon binding to Asf1 and adds a beta strand to the Asf1 beta sheet sandwich. Interactions with both H3 and H4 were required for Asf1 histone chaperone function in vivo and in vitro. The Asf1-H3/H4 structure suggests a "strand-capture" mechanism whereby the H4 tail acts as a lever to facilitate chromatin disassembly/assembly that may be used ubiquitously by histone chaperones.
About this Structure
2HUE is a Protein complex structure of sequences from Saccharomyces cerevisiae and Xenopus laevis. Full crystallographic information is available from OCA.
Reference
Structural basis for the histone chaperone activity of Asf1., English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK, Cell. 2006 Nov 3;127(3):495-508. PMID:17081973
Page seeded by OCA on Mon Mar 31 03:35:32 2008