This is a default text for your page Jennifer Taylor/Sandbox 5. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Background
Proteins are one of the four major macromolecules in biology. Present in nearly every living organism, proteins have a diverse set of functions ranging from regulating cell activity to catalyzing reactions. Proteins are primary made up of amino acids in the primary structure, which consists of the amino acid sequence. The amino acids are set in a particular order so that they perform a specific function, and this order is determined by the DNA gene that codes for amino acids. The secondary structure consists of and . These two structures are helpful in finding similar proteins that perform similar functions. The tertiary and and quaternary structures consist of the specific folding of the complete protein structure, which all comes togther in great importance when classifying proteins. [1]
Enzymes, proteins that catalyze reactions, are grouped into seven major classes based on amino acid sequence similarity and secondary structure proportions. Proteins in each class share unique properties that can classify them into more detailed subclasses. Protein structure and protein function are closely related. This means that identifying highly conserved sequences between two proteins increases the likelihood of discovering shared functions. In this study we attempted to compare the sequence and structure of an uncharacterized protein to that of a protein with a known function in order to understand the former protein’s function. In 2000, the Protein Structure Initiative began an attempt to solve the 3D-structures of proteins with known sequences in order to begin understanding their functions. But, in 2015, the Initiative no longer had the proper funding and stopped, successfully solving 6920 structures, but leaving many structures found without their functions classified as well. What we set out to do was choose a protein with a found structure, perform sequential, structural, and enzymatic analysis.
Bacterial Transformation, Protein Expression, and Protein Purification
First, we needed grow our protein that was inside a plasmid. The NEB bacterial transformation protocol was performed using DH5∂ E.Coli cells in order to insert the plasmid inside the bacteria cells. Next, an overnight culture was performed using to grow more bacteria, and therefore more proteins inside of the DNA. The plasmid was then purified and isolated from genomic DNA, proteins, ribosomes, and cell walls of bacteria using the Zippy miniprep protocol. After determining that T7 polymerase was the polymerase that transcribes 2QRU by computational analysis, it was necessary to use BL21(DE3) E.Coli to transform the cells because DH5∂ does not produce T7 polymerase. We then ran The NEB transformation protocol using BL21 cells and then a liquid culture was grown. With properly grown bacteria, we used the Zippy miniprep protocol was then run to measure the concentration of the plasmid. Next, we needed to determine that our plasmid successfully, purified, so we ran a SDS Page Gel that showed us the weight of our plasmid, that we previously were given by Snao Gene.
Structural Highlights
In our research to find similar proven protein structures to our 2QRU because we know that structure dictates function, we used several computational analysis systems to align our sequences and also align our structures. After obtaining the protein, BLAST, PDB, ProMol were used to find the most structurally similar proteins to 2QRU. First, proteins with similar sequences were searched for, because we assumed that if the sequences were similar, than the structures might be as well. On these databases, 1TAH, 1C4X and 3FAK, all in the esterase sub-family, proved to be the most structurally similar after searching for the the motifs and aligned active sites on ProMol. ProMol was important because we were able to find the . The active site is where reactions happen, so if the active sites were similar, then maybe the reactions would be so too. We used PyMol to align the 3D structures and the active sites to analyze the similarities between the three proteins with known functions. PyMol allowed us to come to the conclusion that since the three proteins were all in the Esterase family, we could then use an assay that would wither prove or disprove if 2QRU was an esterase.
Functional Assay
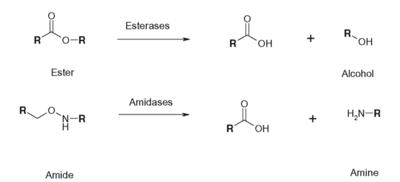
Figure 1: Esterase Reaction
An esterase activity assay using p-Nitrophenyl butyrate and a colorimeter was found online and performed with the elution samples from the Thermo Fisher Protein Purification Assay (3). Besides from the NPB with n-Heptane and the protein, a Tris (hydroxymethyl) aminomethane buffer with a pH of 8 and with 0.01% Triton was made. A blank sample with 1 mL of Tris Buffer and 6.7 uL of varied concentrations of NPB n-Heptane was vortex for 10 seconds and the put into a cuvette and blanked in the colorimeter. For every new concentration, a new blank was made. The concentrations of NPB dissolved in n-Heptane included 0.5M, 0.375M, 0.25M, 0.16M, 0.15M, 0.015M, and 0.075M. The control included 1 mL of Tris Buffer and 6.7 uL of NPB n-Heptane, which was vortexed, and then 6.7 uL of the elution sample from the Thermo Fisher Protein Purification Assay was added, transferred into a cuvette, pipetted up and down, and then the values were recorded every 30 seconds for 2 minutes.
[Image:PNBReaction.png|thumb|left|400px|Figure 2: P-Nitrophenyl Reaction with Lipase]]
Future Directions
The esterase assay proved to be successful in characterizing 2QRU as a esterase. The increasing concentration value indicated that a reaction occurred inside the cuvette containing 2QRU and p-nitrophenyl butyrate. Because p-nitrophenyl butyrate is a substrate that has specific binding for esterases, we can assume that 2QRU may be esterase because a reaction took place inside the cuvette, but more data is needed to make a confident claim. This reaction was not only evident through the OD readings but also through visible observation. Before 2QRU was added to the cuvette, the solution was a cloudy white color. However, once we added the 2QRU there was a visible color change to a bright yellow, confirming that a reaction was taking place within the cuvette.
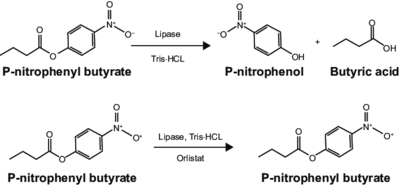
Figure 2: P-Nitrophenyl Reaction with Lipase
After coming to the conclusion that 2QRU is an esterase, our next research activity would be to test if 2QRU is also a lipase. Three students from The Pingry School in New Jersey performed a lipase assay with 2QRU using nitrophenyl palmitate rather than p-nitrophenyl butyrate to determine if 2QRU is a lipase. Nitrophenyl palmitate has (CH2)14 side chain than nitrophenyl butyrate, so the protein has to act on a bigger substrate. If this assay works for us, then would be able to conclude that 2QRU is also a lipase. Another avenue to explore after confirming 2QRU is a lipase is mutagenesis. If we mutated the catalytic triad responsible for the reaction with nitrophenyl palmitate and ran the lipase assay again, we should see no reaction occur in the cuvette. We can use this to confirm that 2QRU is a lipase. The same thing can be done to confirm 2QRU is an esterase if we run the esterase assay using p-nitrophenyl butyrate.
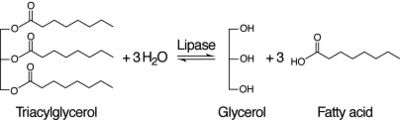
Figure 3: Lipase Reaction



