This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jennifer Taylor/Sandbox 5
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
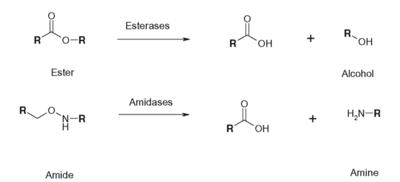
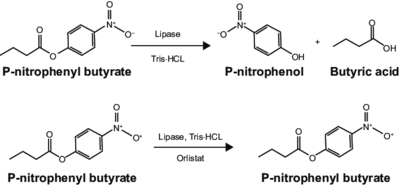
An esterase activity assay using p-Nitrophenyl butyrate and a colorimeter was found online and performed with the elution samples from the Thermo Fisher Protein Purification Assay (3). Besides from the NPB with n-Heptane and the protein, a Tris (hydroxymethyl) aminomethane buffer with a pH of 8 and with 0.01% Triton was made. A blank sample with 1 mL of Tris Buffer and 6.7 uL of varied concentrations of NPB n-Heptane was vortex for 10 seconds and the put into a cuvette and blanked in the colorimeter. For every new concentration, a new blank was made. The concentrations of NPB dissolved in n-Heptane included 0.5M, 0.375M, 0.25M, 0.16M, 0.15M, 0.015M, and 0.075M. The control included 1 mL of Tris Buffer and 6.7 uL of NPB n-Heptane, which was vortexed, and then 6.7 uL of the elution sample from the Thermo Fisher Protein Purification Assay was added, transferred into a cuvette, pipetted up and down, and then the values were recorded every 30 seconds for 2 minutes. | An esterase activity assay using p-Nitrophenyl butyrate and a colorimeter was found online and performed with the elution samples from the Thermo Fisher Protein Purification Assay (3). Besides from the NPB with n-Heptane and the protein, a Tris (hydroxymethyl) aminomethane buffer with a pH of 8 and with 0.01% Triton was made. A blank sample with 1 mL of Tris Buffer and 6.7 uL of varied concentrations of NPB n-Heptane was vortex for 10 seconds and the put into a cuvette and blanked in the colorimeter. For every new concentration, a new blank was made. The concentrations of NPB dissolved in n-Heptane included 0.5M, 0.375M, 0.25M, 0.16M, 0.15M, 0.015M, and 0.075M. The control included 1 mL of Tris Buffer and 6.7 uL of NPB n-Heptane, which was vortexed, and then 6.7 uL of the elution sample from the Thermo Fisher Protein Purification Assay was added, transferred into a cuvette, pipetted up and down, and then the values were recorded every 30 seconds for 2 minutes. | ||
| - | [Image:PNBReaction.png|thumb|left|400px|Figure 2: P-Nitrophenyl Reaction with Lipase]] | + | [[Image:PNBReaction.png|thumb|left|400px|Figure 2: P-Nitrophenyl Reaction with Lipase]] |
==Future Directions== | ==Future Directions== | ||
The esterase assay proved to be successful in characterizing 2QRU as a esterase. The increasing concentration value indicated that a reaction occurred inside the cuvette containing 2QRU and p-nitrophenyl butyrate. Because p-nitrophenyl butyrate is a substrate that has specific binding for esterases, we can assume that 2QRU may be esterase because a reaction took place inside the cuvette, but more data is needed to make a confident claim. This reaction was not only evident through the OD readings but also through visible observation. Before 2QRU was added to the cuvette, the solution was a cloudy white color. However, once we added the 2QRU there was a visible color change to a bright yellow, confirming that a reaction was taking place within the cuvette. | The esterase assay proved to be successful in characterizing 2QRU as a esterase. The increasing concentration value indicated that a reaction occurred inside the cuvette containing 2QRU and p-nitrophenyl butyrate. Because p-nitrophenyl butyrate is a substrate that has specific binding for esterases, we can assume that 2QRU may be esterase because a reaction took place inside the cuvette, but more data is needed to make a confident claim. This reaction was not only evident through the OD readings but also through visible observation. Before 2QRU was added to the cuvette, the solution was a cloudy white color. However, once we added the 2QRU there was a visible color change to a bright yellow, confirming that a reaction was taking place within the cuvette. | ||
| - | |||
| - | [[Image:PNBReaction.png|thumb|left|400px|Figure 2: P-Nitrophenyl Reaction with Lipase]] | ||
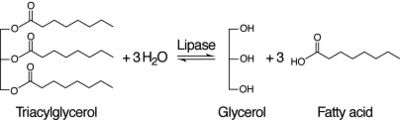
After coming to the conclusion that 2QRU is an esterase, our next research activity would be to test if 2QRU is also a lipase. Three students from The Pingry School in New Jersey performed a lipase assay with 2QRU using nitrophenyl palmitate rather than p-nitrophenyl butyrate to determine if 2QRU is a lipase. Nitrophenyl palmitate has (CH2)14 side chain than nitrophenyl butyrate, so the protein has to act on a bigger substrate. If this assay works for us, then would be able to conclude that 2QRU is also a lipase. Another avenue to explore after confirming 2QRU is a lipase is mutagenesis. If we mutated the catalytic triad responsible for the reaction with nitrophenyl palmitate and ran the lipase assay again, we should see no reaction occur in the cuvette. We can use this to confirm that 2QRU is a lipase. The same thing can be done to confirm 2QRU is an esterase if we run the esterase assay using p-nitrophenyl butyrate. | After coming to the conclusion that 2QRU is an esterase, our next research activity would be to test if 2QRU is also a lipase. Three students from The Pingry School in New Jersey performed a lipase assay with 2QRU using nitrophenyl palmitate rather than p-nitrophenyl butyrate to determine if 2QRU is a lipase. Nitrophenyl palmitate has (CH2)14 side chain than nitrophenyl butyrate, so the protein has to act on a bigger substrate. If this assay works for us, then would be able to conclude that 2QRU is also a lipase. Another avenue to explore after confirming 2QRU is a lipase is mutagenesis. If we mutated the catalytic triad responsible for the reaction with nitrophenyl palmitate and ran the lipase assay again, we should see no reaction occur in the cuvette. We can use this to confirm that 2QRU is a lipase. The same thing can be done to confirm 2QRU is an esterase if we run the esterase assay using p-nitrophenyl butyrate. | ||
Revision as of 13:48, 21 May 2018
2QRU
Here is a cartoon image of my protein:
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644