This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jennifer Taylor/Sandbox 5
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
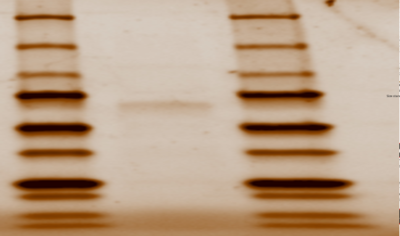
First, we needed grow our protein that was inside a plasmid. The NEB bacterial transformation protocol was performed using DH5∂ E.Coli cells in order to insert the plasmid inside the bacteria cells. Next, an overnight culture was performed using to grow more bacteria, and therefore more proteins inside of the DNA. The plasmid was then purified and isolated from genomic DNA, proteins, ribosomes, and cell walls of bacteria using the Zippy miniprep protocol. After determining that T7 polymerase was the polymerase that transcribes 2QRU by computational analysis, it was necessary to use BL21(DE3) E.Coli to transform the cells because DH5∂ does not produce T7 polymerase. We then ran The NEB transformation protocol using BL21 cells and then a liquid culture was grown. With properly grown bacteria, we used the Zippy miniprep protocol was then run to measure the concentration of the plasmid. Next, we needed to determine that our plasmid successfully, purified, so we ran a SDS Page Gel that showed us the weight of our plasmid, that we previously were given by Snao Gene. The Thermo Fisher Scientific Protein Purification Protocol was used to purify the protein using a His-Pur Nickel-NTA spin column and an equilibrium, a wash, and an elution buffer that we made in class. Centrifuging the column with the elution buffer, after equilibrating and washing the column, allowed protein to flow through, and give us a 2QRU sample. An SDS page gel was used to determine if the final elution samples contained the protein. The protein was successfully expressed and ready to use because its weight on the gel matched 2QRU’s weight found on SnapGene. | First, we needed grow our protein that was inside a plasmid. The NEB bacterial transformation protocol was performed using DH5∂ E.Coli cells in order to insert the plasmid inside the bacteria cells. Next, an overnight culture was performed using to grow more bacteria, and therefore more proteins inside of the DNA. The plasmid was then purified and isolated from genomic DNA, proteins, ribosomes, and cell walls of bacteria using the Zippy miniprep protocol. After determining that T7 polymerase was the polymerase that transcribes 2QRU by computational analysis, it was necessary to use BL21(DE3) E.Coli to transform the cells because DH5∂ does not produce T7 polymerase. We then ran The NEB transformation protocol using BL21 cells and then a liquid culture was grown. With properly grown bacteria, we used the Zippy miniprep protocol was then run to measure the concentration of the plasmid. Next, we needed to determine that our plasmid successfully, purified, so we ran a SDS Page Gel that showed us the weight of our plasmid, that we previously were given by Snao Gene. The Thermo Fisher Scientific Protein Purification Protocol was used to purify the protein using a His-Pur Nickel-NTA spin column and an equilibrium, a wash, and an elution buffer that we made in class. Centrifuging the column with the elution buffer, after equilibrating and washing the column, allowed protein to flow through, and give us a 2QRU sample. An SDS page gel was used to determine if the final elution samples contained the protein. The protein was successfully expressed and ready to use because its weight on the gel matched 2QRU’s weight found on SnapGene. | ||
| - | [[ | + | [[Image:Gel.png|thumb|400 px|Figure 1: SDS Page Gel of 2QRU Purified Protein from Elution Sample]] |
==Structural Highlights== | ==Structural Highlights== | ||
Revision as of 14:28, 21 May 2018
2QRU
Here is a cartoon image of my protein:
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644