This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jennifer Taylor/Sandbox 5
From Proteopedia
| Line 5: | Line 5: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | ==Background== | + | ==Background and Goal== |
Proteins are one of the four major macromolecules in biology. Present in nearly every living organism, proteins have a diverse set of functions ranging from regulating cell activity to catalyzing reactions. Proteins are primary made up of amino acids in the primary structure, which consists of the amino acid sequence. The amino acids are set in a particular order so that they perform a specific function, and this order is determined by the DNA gene that codes for amino acids. The secondary structure consists of <scene name='78/787196/Alpha_helixes_of_2qru/1'>Alpha Helix</scene> and <scene name='78/787196/Beta_sheets_of_2qru/1'>Beta Sheet</scene> . These two structures are helpful in finding similar proteins that perform similar functions. The tertiary and and quaternary structures consist of the specific folding of the complete protein structure, which all comes togther in great importance when classifying proteins. [https://www.khanacademy.org/science/biology/macromolecules/proteins-and-amino-acids/a/orders-of-protein-structure] | Proteins are one of the four major macromolecules in biology. Present in nearly every living organism, proteins have a diverse set of functions ranging from regulating cell activity to catalyzing reactions. Proteins are primary made up of amino acids in the primary structure, which consists of the amino acid sequence. The amino acids are set in a particular order so that they perform a specific function, and this order is determined by the DNA gene that codes for amino acids. The secondary structure consists of <scene name='78/787196/Alpha_helixes_of_2qru/1'>Alpha Helix</scene> and <scene name='78/787196/Beta_sheets_of_2qru/1'>Beta Sheet</scene> . These two structures are helpful in finding similar proteins that perform similar functions. The tertiary and and quaternary structures consist of the specific folding of the complete protein structure, which all comes togther in great importance when classifying proteins. [https://www.khanacademy.org/science/biology/macromolecules/proteins-and-amino-acids/a/orders-of-protein-structure] | ||
Revision as of 00:26, 23 May 2018
2QRU
Here is a cartoon image of my protein:
| |||||||||||
References
1. Alberts, Bruce. Molecular Biology of the Cell. 6th ed. New York, NY: Garland Science, Taylor and Francis Group, 2015.
2. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410. PubMed.
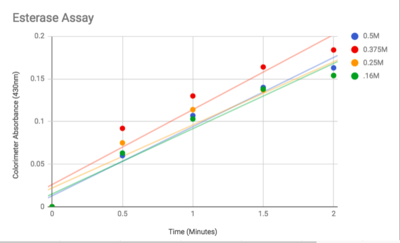
3. "Enzymatic Assay of an Esterase." Sigma-Aldrich. Accessed April 27, 2018. https://www.sigmaaldrich.com/technical-documents/protocols/biology/enzymatic-assay-of-esterase.html.
4. Liisa Holm; Laura M. Laakso (2016) Dali server update. Nucleic acids research 44 (W1), W351-W355. PDF.
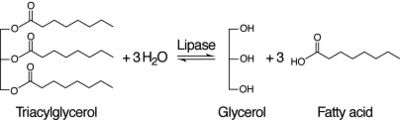
5. Messaoudi, Abdelmonem & Belguith, Hatem & Gram, Imen & Ben Hamida, Jeannette. (2010). Classification of EC 3.1.1.3 bacterial true lipases using phylogenetic analysis. African Journal of Biotechnology. 9. 8243-8247. 10.5897/AJB10.721.
6. "Milestones Tables." PSI Structural Biology Knowledgebase. Accessed April 27, 2018. http://targetdb.pdb.org/Metrics/MilestonesTables.html.
7. Nam K.H., Kim M.Y., Kim S.J., Priyadarshi A., Lee W.H., Hwang K.Y. Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. (2009) Biochem Biophys Res Commun 379: 553-6 Pubmed Article: 19116143
8. Noble M.E., Cleasby A., Johnson L.N., Egmond M.R., Frenken L.G. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
(1993) FEBS Lett 331: 123-8
Pubmed Article: 8405390
9. Nandhagopal, N., Senda, T., Hatta, T., Yamada, A., Masai, E., Fukuda, M., Mitsui, Y. Three-Dimensional Structure of Microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic Acid (Hpda) Hydrolase (Bphd Enzyme) from Rhodococcus Sp. Strain Rha1, in the Pcb Degradation Pathway. (1997) Proc.Jpn.Acad.,Ser.B 73: 154
6. PDB: 2QRU Cuff, M.E., Volkart, L., Moy, S., Joachimiak, A. Structure of an alpha/beta hydrolase superfamily protein from Enterococcus faecalis.
10. ProMol Project. Dr. Paul Craig, Dr. Herbert Bernstein, Dr. Jeff Mills at Rochester Institute of Technology.
11. SnapGene Software (from GSL Biotech; available at snapgene.com).
12. The Pfam protein families database: towards a more sustainable future: R.D. Finn, P. Coggill, R.Y. Eberhardt, S.R. Eddy, J. Mistry, A.L. Mitchell, S.C. Potter, M. Punta, M. Qureshi, A. Sangrador-Vegas, G.A. Salazar, J. Tate, A. BatemanNucleic Acids Research (2016) Database Issue 44:D279-D285
13. The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
15. https://www.livescience.com/45145-how-do-enzymes-work.html