Background
Proteins are an important type of macromolecule in biological systems and can be considered a sequence of subunits known as amino acids. The development of high-throughput genome squencing techniques allowed proteins to be sequenced more quickly than their structures could be solved. In an effort to close this gap, in 2000, the National Institutes of Health launched the 15-year Protein Structure Initiative. Many structures were deposited in the Protein Data Bank, but many of these proteins with solved structures, such as YxiM (PDB ID: 2O14), remain functionally uncharacterized. YxiM is transcribed by the yxiM gene from Bacillus subtilis, a ubiquitous bacterial species that dwells in soil and gastrointestinal tracts. is 375 amino acids in length and its molecular weight is 41.8 kDa. It appears to have two domains: , and .
In silico Analysis
A common theme in biology is that form follows function. Thus, we used computer programs to find which proteins were most homologous to YxiM in terms of sequence and structure, with the expectation that YxiM is likely to be functionally similar to those proteins that have similar sequences and structures.
We used BLAST and PFam to find characterized proteins whose sequences aligned best with YxiM. Sequence analysis suggests that YxiM is a GDSL-like lipase, a type of esterase. Esterases are molecules that hydrolyze (decompose) a class of organic molecules known as esters. GDSL-like lipases demonstrate broad substrate specificity due to their flexible structures. BLAST showed that the proteins 1J00, 1IVN, and 1JRL have the highest sequence homology to YxiM. These proteins are multifunctional hydrolases that show both esterase and protease activity.
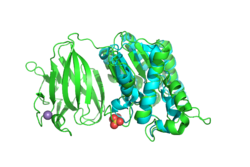
Figure 2: insert your caption
Next, we used PyMOL to align the 3D structures of the BLAST hits with that of YxiM. The proteins 1J00, 1IVN, and 1JRL all align well with the α-helix domain of YxiM.
The Dali server finds the most similar proteins based on 3D structures, and the top 30 hits for YxiM were are all rhamnogalacturonan acetylesterases, GDSL lipases, LAE5s (hydrolases), or acetyl xylan esterases, which further suggests that YxiM is an esterase.
Finally, we used ProMOL to perform a structural alignment of active sites of other proteins with YxiM to predict the active site of YxiM. We found that YxiM aligns best with the active site of IBWR, which is an esterase. The of YxiM consists of amino acids S171, D339, and H342.
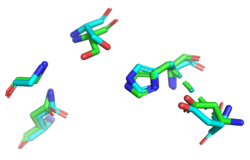
Figure 2: insert your caption
Based on these analyses, we predicted that YxiM is an esterase and proceeded to perform in vitro assays to confirm esterase activity.
Plasmid Purification
In order to study the protein YxiM, we ordered a plasmid that contains the gene that transcribes the protein. A plasmid is a type of circular bacterial DNA. By transforming (inserting) this plasmid (pET21-yxiM) into the bacteria (DH5α Competent E. coli), we can use the bacteria to create more of the plasmid. Then, we performed a DNA miniprep to purify the plasmid for later use.
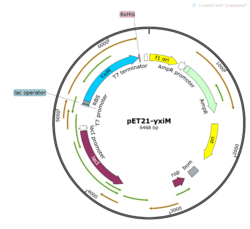
Figure 1: insert your caption
Bacterial Transformation
While DH5α E. coli are good for purifying plasmids, BL21(DE3) E. coli are more efficient for expressing protein. Thus, we transformed the plasmid into BL21(DE3) for the purposes of protein expression. We plated the bacteria on agar with the antibiotic ampicillin. While normal E. coli will die in the presence of ampicillin, the pET21-yxiM plasmid has a gene that allows bacteria to become ampicillin resistant. Thus, only bacteria that were successfully transformed by the plasmid will survive on the ampicillin plate, allowing us to select for bacteria that have been transformed and thus bacteria that will express the protein YxiM.
Protein Expression
After a day, colonies of transformed bacteria were visible on the agar plates. To express YxiM, we inoculated a single colony of bacteria into a liquid culture. In the plasmid, the yxiM gene is under control of the lac operon. This means that in the absence of an inducer, the transcription of the yxiM gene is repressed. Thus, we added IPTG, to activate the operon and drive the transcription of the protein YxiM.
Protein Purification
After several hours, the bacteria have produced a relatively large amount of YxiM. To collect the protein, which at this point remained inside the cells, the bacteria were lysed (their cell walls were burst). The resulting mixture consisted of various cellular proteins and debris. The plasmid DNA sequence that coded for YxiM added a "tag" of histidines at the end of the protein. This allows us to separate YxiM proteins from the other types of proteins in the E. coli cells because when the mixture is passed through a nickel column, the tagged YxiM proteins stick to the column, while the other proteins flow through. Finally, we added elution buffer to the columns, which caused the proteins to detach from the nickel columns, creating a solution of just the YxiM proteins.
Esterase Activity Assay
Now that we had purified protein, we could test the function of YxiM in vitro. Since we believed that YxiM was an ester, we placed it in a buffered solution with 4-nitrophenyl butyrate, a type of ester. Esterases should hydrolyze 4-nitrophenyl butyrate, causing the products butyric acid and 4-nitrophenol to form. Since 4-nitrophenyl is a yellow color, the absorbance of the solution changes as more products are formed. We used colorimeter to measure the absorbance at 430 nm as a proxy for esterase activity. We found that the absorbance increases over time, which suggests that YxiM is indeed an esterase.
Specifically, we found that the Lineweaver-Burk plot of esterase activity is linear. This is typical of enzymes, as predicted by the Michaelis-Menten model of enzyme kinetics. Technically, we did not construct a true Lineweaver-Burk plot, as we used absorbance as a proxy for molar concentration, but absorbance varies linearly with concentration, as shown by the Beer-Lambert law.
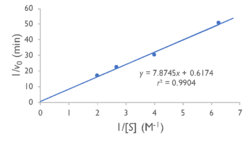
Figure 2: insert your caption
Discussion
YxiM is a previously uncharacterized protein whose crystal structure has been solved and deposited in the PDB.
Protein sequence analysis with BLAST shows that YxiM is likely an esterase. PyMOL shows that the top BLAST hits also align well with the 3D structure of the α-helix domain of YxiM. Almost all the top structural hits in Dali are esterases as well, and ProMOL shows that the active site of YxiM most resembles one of an esterase or protease. The same catalytic triad (S171, D339, H342) is implicated in both protease and esterase activity, suggesting YxiM could be a multifunctional hydrolase. The catalytic motif of the esterase 1BWR aligns particularly well with YxiM.
We tested YxiM for esterase activity in vitro in an effort to confirm the in silico predictions. YxiM showed esterase activity on 4-nitrophenyl butyrate, as absorbance increased during the assay. The Lineweaver-Burk plot of YxiM esterase activity is linear, which is typical of enzymes.
Thus, on the basis of protein sequence and structural analysis in silico and functional assays in vitro, we conclude that YxiM is an esterase.
Future Directions
To further confirm the activity of YxiM as an esterase, we can perform mutagenesis on the putative catalytic triad by performing PCR on the plasmid DNA with specialized primers. If we mutate the catalytic triad, then we expect that the protein will not be able to perform its function anymore. Through another round of transformation and purification of this mutated DNA, we would expect the protein to show no activity in our esterase assay.
Our analysis suggests that YxiM could also demonstrate protease activity. To test this, we could perform protease assays as well.
To further study enzyme kinetics, we could relate absorbance with concentration of protein. We can achieve this by performing a Bradford protein assay to compute the extinction coefficient.




