We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Glycogen Phosphorylase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
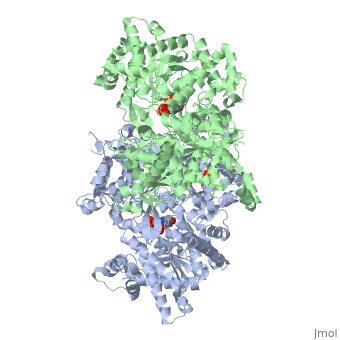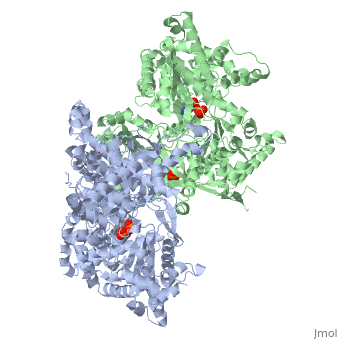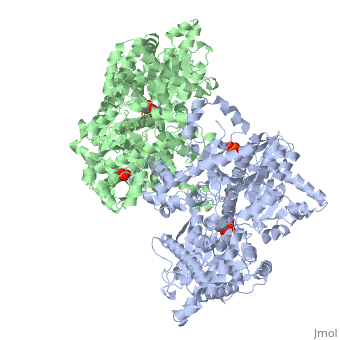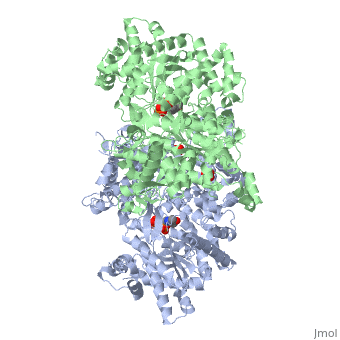
<StructureSection load='1ygp' size='350' side='right' caption='Yeast glycogen phosphorylase dimer with pyridoxal-5-phosphate and phosphate (PDB entry [[1ygp]])' scene=''> | <StructureSection load='1ygp' size='350' side='right' caption='Yeast glycogen phosphorylase dimer with pyridoxal-5-phosphate and phosphate (PDB entry [[1ygp]])' scene=''> | ||
=Introduction= | =Introduction= | ||
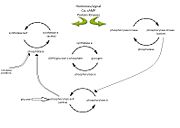
| - | '''Glycogen phosphorylase''' catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and shortened glycogen molecule and is considered the rate limiting step in the degradation of glycogen<ref name="gp">PMID: 15214781 </ref>. It is a part of the glucosyltransferase family and acts on the α-1,4-glycosidic linkage; the phosphorylase comes to a standstill 4 residues from an α-1,6-branchpoint, where debranching enzyme takes over <ref name =“gp3”> PMID: 11949930</ref>. The glucose-1-phophate is then further degraded via the pathway of glycolysis. Studies have found that mammals have liver, muscle and brain isoforms of phosphorylase but it is found among all species; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood <ref name =“gp3”/><ref name="PLP">Palm D, Klein HW, Schinzel R, Buehner M, Helmreich EJM. The role of pyridoxal 5’-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990 Feb 6; 29(5):1099-1107.</ref>. See also [[Glycogen Metabolism & Gluconeogenesis]]. | + | '''Glycogen phosphorylase''' (GP) catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and shortened glycogen molecule and is considered the rate limiting step in the degradation of glycogen<ref name="gp">PMID: 15214781 </ref>. It is a part of the glucosyltransferase family and acts on the α-1,4-glycosidic linkage; the phosphorylase comes to a standstill 4 residues from an α-1,6-branchpoint, where debranching enzyme takes over <ref name =“gp3”> PMID: 11949930</ref>. The glucose-1-phophate is then further degraded via the pathway of glycolysis. Studies have found that mammals have liver, muscle and brain isoforms of phosphorylase but it is found among all species; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood <ref name =“gp3”/><ref name="PLP">Palm D, Klein HW, Schinzel R, Buehner M, Helmreich EJM. The role of pyridoxal 5’-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990 Feb 6; 29(5):1099-1107.</ref>. See also [[Glycogen Metabolism & Gluconeogenesis]]. |
| + | |||
| + | '''GP A''' which is usually active is phosphorylated on Ser 14 of each subunit. GP A is the liver isozyme. '''GP B''' is usually inactive and is the muscle isozyme. GP B is also called '''myophosphorylase'''. | ||
=Structure and Function= | =Structure and Function= | ||
| Line 53: | Line 55: | ||
{{#tree:id=OrganizedByTopic|openlevels=0| | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | *Glycogen phosphorylase | + | *Glycogen phosphorylase B |
| - | **[[ | + | **[[1pyg]] – rGP B + AMP + PDP - rabbit<br /> |
| - | **[[3e3o]] – rGP + IMP<br /> | + | **[[7gpb]], [[8gpb]] – rGP B + AMP + PLP <br /> |
| - | **[[2pyd]], [[2gpb]] – rGP + glucose<br /> | + | **[[3e3n]] – rGP B + AMP + PLP derivative<br /> |
| - | **[[3e3l]], [[1c50]], [[1bx3]], [[2gpn]], [[1gpb]] – rGP<br /> | + | **[[9gpb]], [[1gpb]], [[1bx3]] – rGP B + PLP<br /> |
| - | **[[1abb]] – rGP + | + | **[[3e3o]] – rGP B + IMP<br /> |
| - | **[[1z8d]] – hGP | + | **[[2pyd]], [[2gpb]] – rGP B + glucose<br /> |
| + | **[[3e3l]], [[1c50]], [[1bx3]], [[2gpn]], [[1gpb]] – rGP B + PLP derivative<br /> | ||
| + | **[[1abb]] – rGP B + IMP + PDP<br /> | ||
| + | **[[1z8d]] – hGP B + AMP + glucose - human<br /> | ||
**[[5iko]] – hGP brain form + PLP<br /> | **[[5iko]] – hGP brain form + PLP<br /> | ||
**[[5ikp]] – hGP brain form + PLP + AMP<br /> | **[[5ikp]] – hGP brain form + PLP + AMP<br /> | ||
| - | **[[1fa9]] – hGP liver form + AMP<br /> | ||
**[[1ygp]] – GP + phosphate – yeast | **[[1ygp]] – GP + phosphate – yeast | ||
| - | *Glycogen phosphorylase binary complex with inhibitor | + | *Glycogen phosphorylase B binary complex with inhibitor |
| - | **[[3np7]], [[3np9]], [[3npa]], [[3msc]], [[3mqf]], [[3mrt]], [[3mrv]], [[3mrx]], [[3ms2]], [[3ms4]], [[3ms7]], [[3mt7]], [[3mt8]], [[3mt9]], [[3mta]], [[3mtb]], [[3mtd]], [[3nc4]], [[3l79]], [[3l7a]], [[3l7b]], [[3l7c]], [[3l7d]], [[3g2h]], [[3g2i]], [[3g2j]], [[3g2k]], [[3g2l]], [[3cut]], [[3cuu]], [[3cuv]], [[3cuw]], [[3bcs]], [[3bd7]], [[3bd8]], [[3bda]], [[2qn3]], [[2qn7]], [[2qn8]], [[2qn9]], [[2qnb]], [[2qlm]], [[2qln]], [[2fet]], [[2ff5]], [[2ffr]], [[2f3p]], [[2f3q]], [[2f3s]], [[2f3u]], [[1ww2]], [[1ww3]], [[1xkx]], [[1xl0]], [[1xl1]], [[1xc7]], [[1p4g]], [[1p4h]], [[1p4j]], [[1kti]], [[1k06]], [[1k08]], [[1hlf]], [[1fs4]], [[1ftq]], [[1ftw]], [[1fty]], [[1fu4]], [[1fu7]], [[1fu8]], [[1ggn]], [[2prj]], [[5gpb]], [[3s0j]], [[3sym]], [[3syr]], [[3t3d]], [[3t3e]], [[3t3g]], [[3t3h]], [[3t3i]], [[4ej2]], [[4eke]], [[4eky]], [[4el0]], [[4el5]] – rGP + glucopyranosyl derivative inhibitor<br /> | + | **[[3np7]], [[3np9]], [[3npa]], [[3msc]], [[3mqf]], [[3mrt]], [[3mrv]], [[3mrx]], [[3ms2]], [[3ms4]], [[3ms7]], [[3mt7]], [[3mt8]], [[3mt9]], [[3mta]], [[3mtb]], [[3mtd]], [[3nc4]], [[3l79]], [[3l7a]], [[3l7b]], [[3l7c]], [[3l7d]], [[3g2h]], [[3g2i]], [[3g2j]], [[3g2k]], [[3g2l]], [[3cut]], [[3cuu]], [[3cuv]], [[3cuw]], [[3bcs]], [[3bd7]], [[3bd8]], [[3bda]], [[2qn3]], [[2qn7]], [[2qn8]], [[2qn9]], [[2qnb]], [[2qlm]], [[2qln]], [[2fet]], [[2ff5]], [[2ffr]], [[2f3p]], [[2f3q]], [[2f3s]], [[2f3u]], [[1ww2]], [[1ww3]], [[1xkx]], [[1xl0]], [[1xl1]], [[1xc7]], [[1p4g]], [[1p4h]], [[1p4j]], [[1kti]], [[1k06]], [[1k08]], [[1hlf]], [[1fs4]], [[1ftq]], [[1ftw]], [[1fty]], [[1fu4]], [[1fu7]], [[1fu8]], [[1ggn]], [[2prj]], [[5gpb]], [[3s0j]], [[3sym]], [[3syr]], [[3t3d]], [[3t3e]], [[3t3g]], [[3t3h]], [[3t3i]], [[4ej2]], [[4eke]], [[4eky]], [[4el0]], [[4el5]] – rGP B + glucopyranosyl derivative inhibitor<br /> |
| - | **[[3g2n]] – rGP + acylglucosylamine<br /> | + | **[[3g2n]] – rGP B + acylglucosylamine<br /> |
| - | **[[3ebo]] – rGP + chrysin<br /> | + | **[[3ebo]] – rGP B + chrysin<br /> |
| - | **[[3ebp]], [[1gfz]], [[1c8k]], [[1e1y]] – rGP + flavopiridol<br /> | + | **[[3ebp]], [[1gfz]], [[1c8k]], [[1e1y]] – rGP B + flavopiridol<br /> |
| - | **[[ | + | **[[2pyi]], [[6f3l]], [[6f3r]], [[6f3s]], [[6f3u]] – rGP B + triazol derivative<br /> |
| - | + | ||
**[[1gg8]] – rGP + glucoside inhibitor<br /> | **[[1gg8]] – rGP + glucoside inhibitor<br /> | ||
| - | **[[2pri]]– rGP + deoxyglucosephosphate inhibitor<br /> | + | **[[2pri]]– rGP B + deoxyglucosephosphate inhibitor<br /> |
| - | **[[3gpb]], [[4gpb]], [[6gpb]] – rGP + phosphorylated glucose derivative | + | **[[3gpb]], [[4gpb]], [[6gpb]] – rGP B + phosphorylated glucose derivative<br /> |
| - | + | **[[3bcr]] – rGP B + AZT<br /> | |
| - | **[[3bcr]] – rGP + AZT<br /> | + | **[[3bcu]] – rGP B + thymidine<br /> |
| - | **[[3bcu]] – rGP + thymidine<br /> | + | **[[2gj4]] – rGP B + ligand<br /> |
| - | **[[2gj4]] – rGP + ligand<br /> | + | **[[1gpy]] – rGP B + glucose-6-phosphate<br /> |
| - | **[[1gpy]] – rGP + glucose-6-phosphate<br /> | + | **[[2gm9]] – rGP B + thienopyrrole<br /> |
| - | **[[2gm9]] – rGP + thienopyrrole<br /> | + | **[[5mem]] – rGP B + pyrimidine derivative inhibitor<br /> |
| - | **[[ | + | **[[2g9q]], [[2g9r]], [[2g9u]], [[2g9v]] – rGP B + iminosugar<br /> |
| - | **[[2g9q]], [[2g9r]], [[2g9u]], [[2g9v]] – rGP + iminosugar<br /> | + | **[[2amv]], [[1c50]], [[1axr]] – rGP B + pyridine derivative inhibitor<br /> |
| - | **[[2amv]] – rGP + pyridine derivative inhibitor<br /> | + | **[[2ieg]], [[2iei]] – rGP B + quinolone derivative inhibitor<br /> |
| - | **[[2ieg]], [[2iei]] – rGP + quinolone derivative inhibitor<br /> | + | **[[1z62]], [[1uzu]] - rGP B + indirubin derivative inhibitor<br /> |
| - | **[[1z62]], [[1uzu]] - rGP + indirubin derivative inhibitor<br /> | + | **[[3bd6]] – rGP B + ribofuranosyl cyanuric acid<br /> |
| - | **[[3bd6]] – rGP + ribofuranosyl cyanuric acid<br /> | + | **[[2qrg]], [[2qrh]], [[2qrm]], [[2qrp]], [[2qrq]], [[4ctm]], [[4ctn]], [[4cto]] – rGP B + spiro-isoxazoline inhibitor<br /> |
| - | **[[2qrg]], [[2qrh]], [[2qrm]], [[2qrp]], [[2qrq]], [[4ctm]], [[4ctn]], [[4cto]] – rGP + spiro-isoxazoline inhibitor<br /> | + | **[[1a8i]] - rGP B + spiro-hydantoine inhibitor<br /> |
| - | **[[1a8i]] - rGP + spiro-hydantoine inhibitor<br /> | + | **[[2qn1]], [[2qn2]] – rGP B + pentacyclic triterpene inhibitor<br /> |
| - | **[[2qn1]], [[2qn2]] – rGP + pentacyclic triterpene inhibitor<br /> | + | **[[2off]] - rGP B + allosteric inhibitor<br /> |
| - | **[[2off | + | **[[1wv0]], [[1wv1]], [[1wuy]] - rGP B + acyl urea derivative inhibitor<br /> |
| - | **[[1wv0]], [[1wv1]], [[1wuy]] - rGP + acyl urea derivative inhibitor<br /> | + | **[[1z6p]], [[1z6q]] - rGP B + AMP site inhibitor<br /> |
| - | **[[1z6p]], [[1z6q]] - rGP + AMP site inhibitor<br /> | + | **[[1b4d]] – rGP B + amidocarbamate inhibitor<br /> |
| - | **[[1b4d]] – rGP + amidocarbamate inhibitor<br /> | + | **[[1p29]], [[1p2b]], [[1p2d]], [[1p2g]] - rGP B + cyclodextrin derivative inhibitor<br /> |
| - | **[[1p29]], [[1p2b]], [[1p2d]], [[1p2g]] - rGP + cyclodextrin derivative inhibitor<br /> | + | |
**[[1lwn]], [[1lwo]] – rGP + hypoglycaemic drug<br /> | **[[1lwn]], [[1lwo]] – rGP + hypoglycaemic drug<br /> | ||
| - | **[[2gpa]], [[2amv]], [[3amv]], [[3zcp]], [[3zcq]], [[3zcr]], [[3zcs]], [[3zct]], [[3zcu]], [[3zcv]], [[4yi3]], [[4yi5]], [[4yua]], [[4z5x]] - rGP | + | **[[2gpa]], [[2amv]], [[3amv]], [[3zcp]], [[3zcq]], [[3zcr]], [[3zcs]], [[3zct]], [[3zcu]], [[3zcv]], [[4yi3]], [[4yi5]], [[4yua]], [[4z5x]] - rGP B + antidiabetic drug<br /> |
| - | **[[5jtt]], [[5jtu]] - rGP | + | **[[5jtt]], [[5jtu]] - rGP B + imidazole derivative<br /> |
| - | **[[ | + | **[[5lre]], [[5lrc]], [[5lrd]], [[5lrf]] - rGP B + triazole derivative<br /> |
| - | **[[4mho]], [[4mhs]], [[4mi3]], [[4mi6]], [[4mi9]], [[4mic]] - rGP + glucopyranosyl derivative<br /> | + | **[[4mho]], [[4mhs]], [[4mi3]], [[4mi6]], [[4mi9]], [[4mic]] - rGP B + glucopyranosyl derivative<br /> |
| - | **[[4mra]] - rGP + quercetin<br /> | + | **[[4mra]] - rGP B + quercetin<br /> |
| - | **[[ | + | **[[5mcb]] – rGP B + chlorogenic acid<br /> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
**[[2c4m]] – GP + vitamin B6 complex – ''Corynebacterium callunae'' | **[[2c4m]] – GP + vitamin B6 complex – ''Corynebacterium callunae'' | ||
| - | *Glycogen phosphorylase | + | *Glycogen phosphorylase B higher complexes |
| + | |||
| + | **[[1h5u]] - rGP B + antidiabetic drug + glucose<br /> | ||
| + | **[[6gpb]] - rGP B + heptulose phosphate + PLP + AMP + glucose<br /> | ||
| + | **[[5o52]], [[5o56]] – rGP B + PLP + glucosaminyl heterocycle inhibitor<br /> | ||
| + | **[[5o50]], [[5o54]] – rGP B + PLP + IMP + glucosaminyl heterocycle inhibitor<br /> | ||
| + | **[[6f3j]], [[5owy]], [[5owz]], [[5ox0]], [[5ox1]], [[5ox3]], [[5ox4]] – rGP B + PLP + IMP + inhibitor<br /> | ||
| + | |||
| + | *Glycogen phosphorylase A | ||
| - | **[[ | + | **[[1gpa]] – rGP A<br /> |
| - | **[[ | + | **[[1fa9]] – hGP A liver form + AMP<br /> |
| - | **[[ | + | **[[3dd1]], [[3dds]], [[3ddw]] – rGP A + anthranilimide<br /> |
| - | **[[2zb2]] – hGP + carboxamide derivative + glucose<br /> | + | **[[3ceh]], [[3cej]], [[3cem]] – hGP A + allosteric inhibitor <br /> |
| - | **[[1em6]], [[1exv]] – hGP + nucleoside + inhibitor<br /> | + | **[[2qll]] – hGP A + GL C terminal peptide<br /> |
| - | **[[1l5q]], [[1l7x]] – hGP + caffeine + glucopyranosyl derivative + inhibitor<br /> | + | **[[2ati]], [[1wut]] – hGP A + acyl urea derivative inhibitor<br /> |
| - | **[[1l5r]] - hGP + riboflavin + glucopyranosyl derivative + inhibitor<br /> | + | **[[1xoi]] - hGP A + indoloyl derivative inhibitor<br /> |
| - | **[[1l5s]] - hGP + uric acid + glucopyranosyl derivative + inhibitor | + | **[[1fc0]] - hGP A + glucopyranosyl derivative<br /> |
| + | **[[2zb2]] – hGP A liver + carboxamide derivative + glucose<br /> | ||
| + | **[[1em6]], [[1exv]] – hGP A liver + nucleoside + inhibitor<br /> | ||
| + | **[[1l5q]], [[1l7x]] – hGP A liver + caffeine + glucopyranosyl derivative + inhibitor<br /> | ||
| + | **[[1l5r]] - hGP A liver + riboflavin + glucopyranosyl derivative + inhibitor<br /> | ||
| + | **[[1l5s]] - hGP A liver + uric acid + glucopyranosyl derivative + inhibitor<br /> | ||
| + | **[[1noi]], [[1noj]], [[1nok]] – rGP A + transition state analog + phosphate<br /> | ||
| + | **[[1c8l]] - rGP A + antidiabetic drug + caffeine<br /> | ||
}} | }} | ||
=Additional Resources= | =Additional Resources= | ||
Revision as of 09:25, 4 June 2018
| |||||||||||
3D structures of glycogen phosphorylase
Updated on 04-June-2018
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ 1.0 1.1 1.2 Kristiansen M, Andersen B, Iversen LF, Westergaard N. Identification, synthesis, and characterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site. J Med Chem. 2004 Jul 1;47(14):3537-45. PMID:15214781 doi:10.1021/jm031121n
- ↑ 2.0 2.1 Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002 Mar;2(2):101-20. PMID:11949930
- ↑ 3.0 3.1 3.2 3.3 Palm D, Klein HW, Schinzel R, Buehner M, Helmreich EJM. The role of pyridoxal 5’-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990 Feb 6; 29(5):1099-1107.
- ↑ 4.0 4.1 4.2 4.3 4.4 Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991 Mar 5;218(1):233-60. PMID:1900534
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Fletterick RJ, Sprang SR. Glycogen phosphorylase Structures and function. Accounts of Chemical Research. 1982 Nov; 15(11):361-369.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 Johnson LH. Glycogen Phosphorylase: Control by phosphorylation and allosteric effectors. The FASEB Journal. 1992 March;6:2274-2282.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Amy Chahal, Ann Taylor, Alexander Berchansky, Joel L. Sussman, Riley Hicks, Andrea Gorrell, David Canner