2j05
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 2j05 |SIZE=350|CAPTION= <scene name='initialview01'>2j05</scene>, resolution 1.50Å | |PDB= 2j05 |SIZE=350|CAPTION= <scene name='initialview01'>2j05</scene>, resolution 1.50Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= | + | |LIGAND= <scene name='pdbligand=MSE:SELENOMETHIONINE'>MSE</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2j05 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2j05 OCA], [http://www.ebi.ac.uk/pdbsum/2j05 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=2j05 RCSB]</span> | ||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
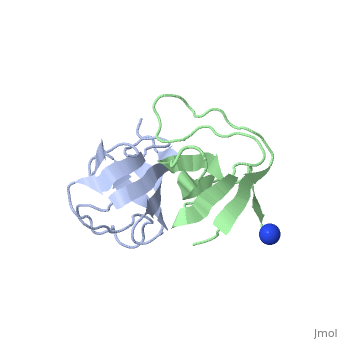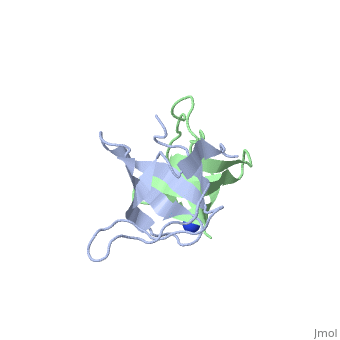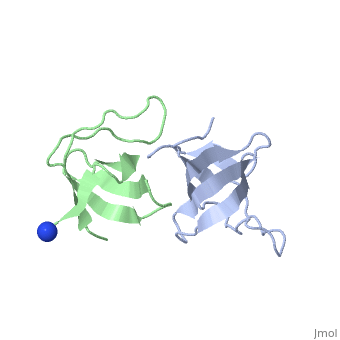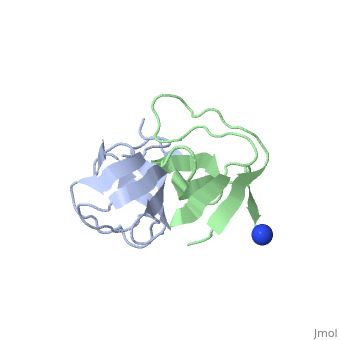
X-ray structures of two crystal forms of the Src homology 3 domain (SH3) of the Ras GTPase activating protein (RasGAP) were determined at 1.5 and 1.8A resolution. The overall structure comprises a single domain with two tightly packed beta-sheets linked by a short helical segment. An important motif for peptide binding in other SH3 domains is not conserved in RasGAP. The RasGAP SH3 domain forms dimers in the crystal structures, which may provide new functional insight. The dimer interface involves residues also present in a peptide previously identified as an apoptotic sensitizer of tumor cells. | X-ray structures of two crystal forms of the Src homology 3 domain (SH3) of the Ras GTPase activating protein (RasGAP) were determined at 1.5 and 1.8A resolution. The overall structure comprises a single domain with two tightly packed beta-sheets linked by a short helical segment. An important motif for peptide binding in other SH3 domains is not conserved in RasGAP. The RasGAP SH3 domain forms dimers in the crystal structures, which may provide new functional insight. The dimer interface involves residues also present in a peptide previously identified as an apoptotic sensitizer of tumor cells. | ||
| - | |||
| - | ==Disease== | ||
| - | Known diseases associated with this structure: Basal cell carcinoma, somatic OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=139150 139150]], Capillary malformation-arteriovenous malformation OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=139150 139150]], Parkes Weber syndrome OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=139150 139150]] | ||
==About this Structure== | ==About this Structure== | ||
| Line 39: | Line 39: | ||
[[Category: src homology 3]] | [[Category: src homology 3]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 03:51:10 2008'' |
Revision as of 00:51, 31 March 2008
| |||||||
| , resolution 1.50Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | |||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
CRYSTAL STRUCTURE OF THE RASGAP SH3 DOMAIN AT 1.5 ANGSTROM RESOLUTION
Overview
X-ray structures of two crystal forms of the Src homology 3 domain (SH3) of the Ras GTPase activating protein (RasGAP) were determined at 1.5 and 1.8A resolution. The overall structure comprises a single domain with two tightly packed beta-sheets linked by a short helical segment. An important motif for peptide binding in other SH3 domains is not conserved in RasGAP. The RasGAP SH3 domain forms dimers in the crystal structures, which may provide new functional insight. The dimer interface involves residues also present in a peptide previously identified as an apoptotic sensitizer of tumor cells.
About this Structure
2J05 is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
High resolution crystal structures of the p120 RasGAP SH3 domain., Ross B, Kristensen O, Favre D, Walicki J, Kastrup JS, Widmann C, Gajhede M, Biochem Biophys Res Commun. 2007 Feb 9;353(2):463-8. Epub 2006 Dec 18. PMID:17188236
Page seeded by OCA on Mon Mar 31 03:51:10 2008