We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Gyrase
From Proteopedia
(Difference between revisions)
| Line 35: | Line 35: | ||
*Gyrase Subunit B | *Gyrase Subunit B | ||
| - | **[[3g75]], [[3g7b]], [[3g7e]], [[3ttz]], [[3u2d]], [[3u2k]], [[4p8o]], [[5cph]], [[5ctu]], [[5ctw]], [[5ctx]], [[5cty]] – GyrB+ inhibitor – ''Staphylococcus aureus''<br /> | + | **[[3g75]], [[3g7b]], [[3g7e]], [[3ttz]], [[3u2d]], [[3u2k]], [[4p8o]], [[5cph]], [[5ctu]], [[5ctw]], [[5ctx]], [[5cty]] – GyrB ATP-binding domain residues 2-234 + inhibitor – ''Staphylococcus aureus''<br /> |
**[[5d6p]], [[5d6q]], [[5d7c]], [[5d7d]], [[5d7r]] - SaGyrB ATP-binding domain + ligand<br /> | **[[5d6p]], [[5d6q]], [[5d7c]], [[5d7d]], [[5d7r]] - SaGyrB ATP-binding domain + ligand<br /> | ||
| + | **[[5npk]], [[5npp]] - SaGyrB residues 410-1491 + DNA<br /> | ||
**[[5mmn]], [[5mmo]], [[5mmp]] - EcGyrB ATP-binding domain + antibacterial<br /> | **[[5mmn]], [[5mmo]], [[5mmp]] - EcGyrB ATP-binding domain + antibacterial<br /> | ||
**[[4urm]], [[4uro]] - SaGyrB N-terminal + antibiotic<br /> | **[[4urm]], [[4uro]] - SaGyrB N-terminal + antibiotic<br /> | ||
| Line 61: | Line 62: | ||
**[[4tma]] - SaGyrB C-terminal- + SaGyrA N-terminal + Gyr inhibitor YACG<br /> | **[[4tma]] - SaGyrB C-terminal- + SaGyrA N-terminal + Gyr inhibitor YACG<br /> | ||
**[[3nuh]] – EcGyrA+EcGyrB<br /> | **[[3nuh]] – EcGyrA+EcGyrB<br /> | ||
| - | **[[5cdn]], [[5cdo]], [[5cdp]] – SaGyrA + SaGyrB + DNA<br /> | + | **[[5cdn]], [[5cdo]], [[5cdp]], [[6fqv]] – SaGyrA + SaGyrB + DNA<br /> |
| - | **[[5cdq]] - SaGyrB + GyrA + DNA + antibiotic<br /> | + | **[[5cdq]], [[6fqm]], [[6fqs]] - SaGyrB + GyrA + DNA + antibiotic<br /> |
**[[5cdm]] – SaGyrA + SaGyrB (mutant) + DNA<br /> | **[[5cdm]] – SaGyrA + SaGyrB (mutant) + DNA<br /> | ||
**[[5cdr]] – SaGyrA (mutant) + SaGyrB (mutant) + DNA<br /> | **[[5cdr]] – SaGyrA (mutant) + SaGyrB (mutant) + DNA<br /> | ||
Revision as of 10:19, 7 June 2018
| |||||||||||
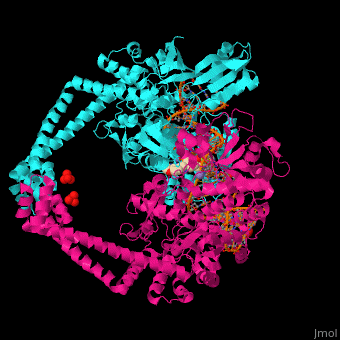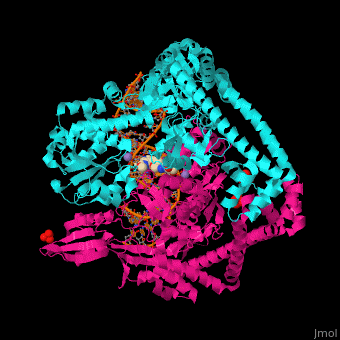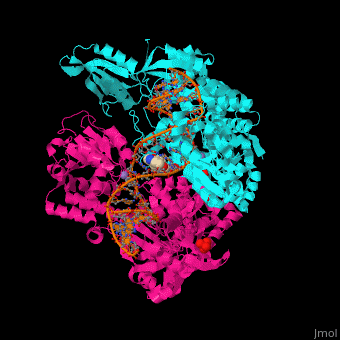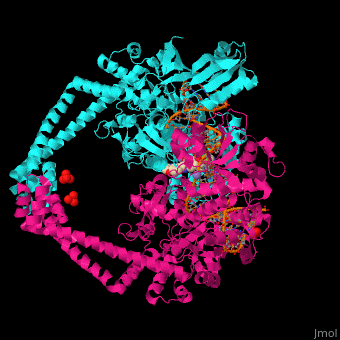
3D Structure of Gyrase
Updated on 07-June-2018
Additional Resources
For additional information, see: Bacterial Infections
References
- ↑ Reece RJ, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26(3-4):335-75. PMID:1657531 doi:http://dx.doi.org/10.3109/10409239109114072
- ↑ Napoli A, Valenti A, Salerno V, Nadal M, Garnier F, Rossi M, Ciaramella M. Functional interaction of reverse gyrase with single-strand binding protein of the archaeon Sulfolobus. Nucleic Acids Res. 2005 Jan 26;33(2):564-76. Print 2005. PMID:15673717 doi:http://dx.doi.org/10.1093/nar/gki202
- ↑ Aubry A, Pan XS, Fisher LM, Jarlier V, Cambau E. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob Agents Chemother. 2004 Apr;48(4):1281-8. PMID:15047530
- ↑ Singh SB, Kaelin DE, Wu J, Miesel L, Tan CM, Meinke PT, Olsen D, Lagrutta A, Bradley P, Lu J, Patel S, Rickert KW, Smith RF, Soisson S, Wei C, Fukuda H, Kishii R, Takei M, Fukuda Y. Oxabicyclooctane-linked novel bacterial topoisomerase inhibitors as broad spectrum antibacterial agents. ACS Med Chem Lett. 2014 Mar 12;5(5):609-14. doi: 10.1021/ml500069w. eCollection, 2014 May 8. PMID:24900889 doi:http://dx.doi.org/10.1021/ml500069w
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman