This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2j92
From Proteopedia
| Line 5: | Line 5: | ||
|SITE= | |SITE= | ||
|LIGAND= | |LIGAND= | ||
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Picornain_3C Picornain 3C], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.22.28 3.4.22.28] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Picornain_3C Picornain 3C], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.22.28 3.4.22.28] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2j92 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2j92 OCA], [http://www.ebi.ac.uk/pdbsum/2j92 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=2j92 RCSB]</span> | ||
}} | }} | ||
| Line 16: | Line 19: | ||
==About this Structure== | ==About this Structure== | ||
| - | 2J92 is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/ | + | 2J92 is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Viruses Viruses]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2J92 OCA]. |
==Reference== | ==Reference== | ||
Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis., Sweeney TR, Roque-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S, J Virol. 2007 Jan;81(1):115-24. Epub 2006 Oct 25. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17065215 17065215] | Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis., Sweeney TR, Roque-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S, J Virol. 2007 Jan;81(1):115-24. Epub 2006 Oct 25. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17065215 17065215] | ||
| - | [[Category: Foot-and-mouth disease virus (strain a10-61)]] | ||
[[Category: Picornain 3C]] | [[Category: Picornain 3C]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
| + | [[Category: Viruses]] | ||
[[Category: Birtley, J R.]] | [[Category: Birtley, J R.]] | ||
[[Category: Curry, S.]] | [[Category: Curry, S.]] | ||
| Line 33: | Line 36: | ||
[[Category: thiol protease]] | [[Category: thiol protease]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 03:54:48 2008'' |
Revision as of 00:54, 31 March 2008
| |||||||
| , resolution 2.20Å | |||||||
|---|---|---|---|---|---|---|---|
| Activity: | Picornain 3C, with EC number 3.4.22.28 | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
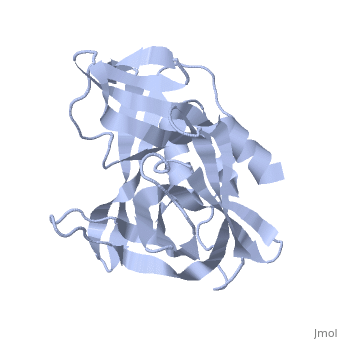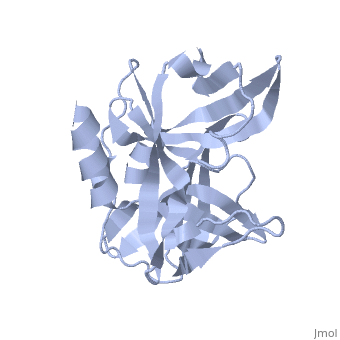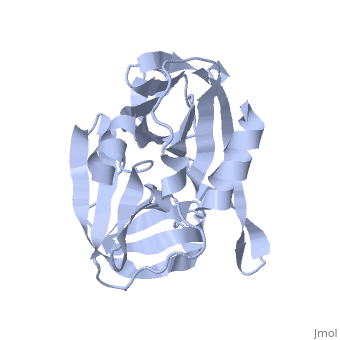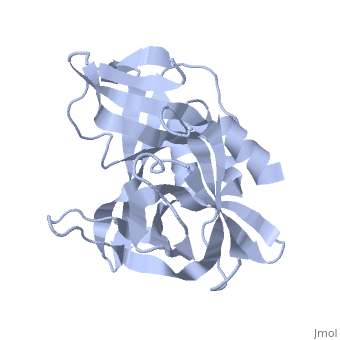
3C PROTEASE FROM TYPE A10(61) FOOT-AND-MOUTH DISEASE VIRUS-CRYSTAL PACKING MUTANT (K51Q)
Overview
The 3C protease (3C(pro)) from foot-and-mouth disease virus (FMDV), the causative agent of a widespread and economically devastating disease of domestic livestock, is a potential target for antiviral drug design. We have determined the structure of a new crystal form of FMDV 3C(pro), a chymotrypsin-like cysteine protease, which reveals features that are important for catalytic activity. In particular, we show that a surface loop which was disordered in previous structures adopts a beta-ribbon structure that is conformationally similar to equivalent regions on other picornaviral 3C proteases and some serine proteases. This beta-ribbon folds over the peptide binding cleft and clearly contributes to substrate recognition. Replacement of Cys142 at the tip of the beta-ribbon with different amino acids has a significant impact on enzyme activity and shows that higher activity is obtained with more hydrophobic side chains. Comparison of the structure of FMDV 3C(pro) with homologous enzyme-peptide complexes suggests that this correlation arises because the side chain of Cys142 contacts the hydrophobic portions of the P2 and P4 residues in the peptide substrate. Collectively, these findings provide compelling evidence for the role of the beta-ribbon in catalytic activity and provide valuable insights for the design of FMDV 3C(pro) inhibitors.
About this Structure
2J92 is a Single protein structure of sequence from Viruses. Full crystallographic information is available from OCA.
Reference
Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis., Sweeney TR, Roque-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S, J Virol. 2007 Jan;81(1):115-24. Epub 2006 Oct 25. PMID:17065215
Page seeded by OCA on Mon Mar 31 03:54:48 2008