We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Ricardo Alberto Chiong Zevallos/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
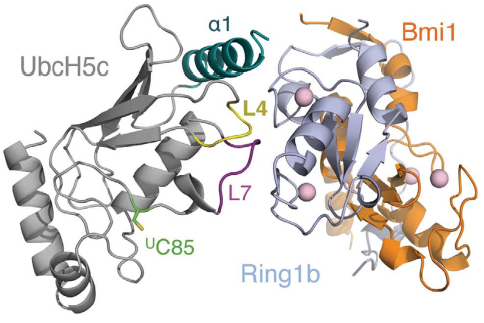
In humans, there are multiple E2 ubiquitin-conjugating enzymes, but only UbcH5 and UbcH6 interact with the BMI1/RING1b heterodimer and promote the monoubiquitination of histone H2A at Lysine 119. The subtype UbcH5c is capable of making more polyubiquitin chains then UbcH5a and UbcH5b, in contrast UbcH6 doesn't make polyubiquitin chains (ref Buchwalld 2006). The BMI1/RING1b heterodimer interface buries a total of 2500 Ų surface area from the two proteins while the UbcH5c/RING1b interface has only 507Ų. The discrepancy between the areas of interface is reflected in the low affinity interaction between BMI1/RING1b and UbcH5c (Buchwald et al, 2006). The RING1b-binding surface on UbcH5c consists of two loops (L4 and L7), as well as residues from the N-terminal a helix (a1). | In humans, there are multiple E2 ubiquitin-conjugating enzymes, but only UbcH5 and UbcH6 interact with the BMI1/RING1b heterodimer and promote the monoubiquitination of histone H2A at Lysine 119. The subtype UbcH5c is capable of making more polyubiquitin chains then UbcH5a and UbcH5b, in contrast UbcH6 doesn't make polyubiquitin chains (ref Buchwalld 2006). The BMI1/RING1b heterodimer interface buries a total of 2500 Ų surface area from the two proteins while the UbcH5c/RING1b interface has only 507Ų. The discrepancy between the areas of interface is reflected in the low affinity interaction between BMI1/RING1b and UbcH5c (Buchwald et al, 2006). The RING1b-binding surface on UbcH5c consists of two loops (L4 and L7), as well as residues from the N-terminal a helix (a1). | ||
[[Image:Interaction 1 between Bmi1.Ring1b and UbcH5c.png]] | [[Image:Interaction 1 between Bmi1.Ring1b and UbcH5c.png]] | ||
| + | |||
Representation of Bmi1/Ring1b-UbcH5c complex structure, with UbcH5c in grey, L4 in yellow, L7 in purple, Ring1b in light blue, and Bmi1 in orange. <ref name="embo">Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297</ref> | Representation of Bmi1/Ring1b-UbcH5c complex structure, with UbcH5c in grey, L4 in yellow, L7 in purple, Ring1b in light blue, and Bmi1 in orange. <ref name="embo">Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297</ref> | ||
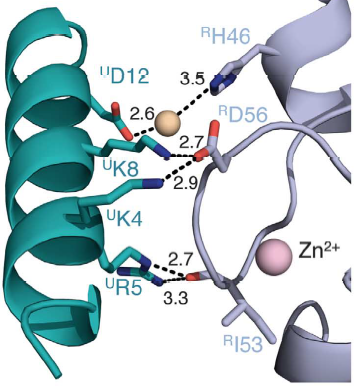
Salt bridges are formed between Lys4 and Lys8 from the a1 helix of UbcH5c (ULys4 and ULys8) with Asp56 on Ring1b (RAsp56) | Salt bridges are formed between Lys4 and Lys8 from the a1 helix of UbcH5c (ULys4 and ULys8) with Asp56 on Ring1b (RAsp56) | ||
| - | [[Image:Interaction 2 between Bmi1.Ring1b and UbcH5c.png | + | [[Image:Interaction 2 between Bmi1.Ring1b and UbcH5c.png]] |
| - | Interactions between Ring1b, in gray, and UbcH5c, in green, along the N-terminal a-helix. The sidechains involved are shown in stick format. Hydrogen-bond distances are given in angstroms. <ref name="embo">Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297</ref>]] | ||
| + | Interactions between Ring1b, in gray, and UbcH5c, in green, along the N-terminal a-helix. The sidechains involved are shown in stick format. Hydrogen-bond distances are given in angstroms. <ref name="embo">Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297</ref> | ||
In the SPA motif of the UbcH5 (L7 residues USer94, UPro95, and UAla96), the sidechain hydroxyl of USer94 makes a hydrogen bond with the backbone carbonyl of RPro88 (from RING1b). Hydrophobic interactions between UPro95 and UAla96 with RIle53 and RPro88 help to stabilize the interaction between UbcH5 and RING1b. | In the SPA motif of the UbcH5 (L7 residues USer94, UPro95, and UAla96), the sidechain hydroxyl of USer94 makes a hydrogen bond with the backbone carbonyl of RPro88 (from RING1b). Hydrophobic interactions between UPro95 and UAla96 with RIle53 and RPro88 help to stabilize the interaction between UbcH5 and RING1b. | ||
| - | + | ||
Canonical PRC1 such as BMI1/RING1b have intrinsically very low enzymatic activity compared with non-canonical PRC1, although there are only subtle differences between the structure of canonical and non-canonical complexes. Two charged helix alpha 3 residues present in a modeled BMI1 (ref taherbhoy 2015) seems responsible for the low activity of BMI1/RING1b, K73 and D77 form a salt bridge that may limit efficient ubiquitin transfer. In a computational modeling, BMI1 K73 clashes sterically with Ubiquitin, which implies that K73 must move to allow Ub to bind in an activated conformation or that Ub adopts a less optimal position during monoubiquitination. Either of these events could provide an energy barrier, slowing transfer catalyzed by BMI1. The intrinsically low activity of the BMI1/RING1b is offset by a relatively favorable interaction between E3–E2-Ub and nucleosome substrate, resulting in a site-specific monoubiquitination efficient enough. The energy barrier may be responsible for increasing the fidelity of the transfer to the appropriate substrate. Also, canonical and non-canonical differ in targeting sub-units, target genomic loci, and genes expression regulation. | Canonical PRC1 such as BMI1/RING1b have intrinsically very low enzymatic activity compared with non-canonical PRC1, although there are only subtle differences between the structure of canonical and non-canonical complexes. Two charged helix alpha 3 residues present in a modeled BMI1 (ref taherbhoy 2015) seems responsible for the low activity of BMI1/RING1b, K73 and D77 form a salt bridge that may limit efficient ubiquitin transfer. In a computational modeling, BMI1 K73 clashes sterically with Ubiquitin, which implies that K73 must move to allow Ub to bind in an activated conformation or that Ub adopts a less optimal position during monoubiquitination. Either of these events could provide an energy barrier, slowing transfer catalyzed by BMI1. The intrinsically low activity of the BMI1/RING1b is offset by a relatively favorable interaction between E3–E2-Ub and nucleosome substrate, resulting in a site-specific monoubiquitination efficient enough. The energy barrier may be responsible for increasing the fidelity of the transfer to the appropriate substrate. Also, canonical and non-canonical differ in targeting sub-units, target genomic loci, and genes expression regulation. | ||
Revision as of 12:48, 17 June 2018
| |||||||||||
References
- ↑ 1.0 1.1 Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297
https://drive.google.com/drive/folders/1l195aNuY6joOd74GKKxa-XWTRMBv_uWF?usp=sharing