This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Ricardo Alberto Chiong Zevallos/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
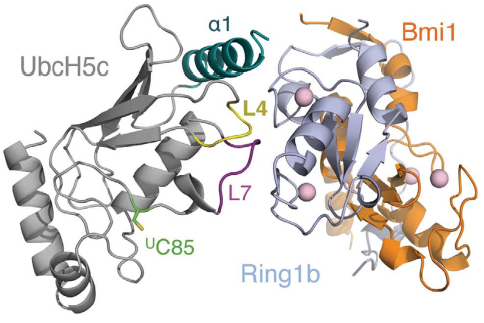
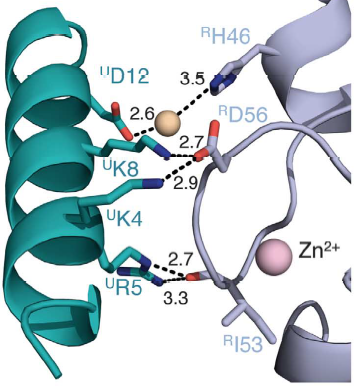
Interactions between Ring1b, in gray, and UbcH5c, in green, along the N-terminal a-helix. The sidechains involved are shown in stick format. Hydrogen-bond distances are given in angstroms. <ref name="embo">Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297</ref> | Interactions between Ring1b, in gray, and UbcH5c, in green, along the N-terminal a-helix. The sidechains involved are shown in stick format. Hydrogen-bond distances are given in angstroms. <ref name="embo">Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297</ref> | ||
| + | |||
In the SPA motif of the UbcH5 (L7 residues USer94, UPro95, and UAla96), the sidechain hydroxyl of USer94 makes a hydrogen bond with the backbone carbonyl of RPro88 (from RING1b). Hydrophobic interactions between UPro95 and UAla96 with RIle53 and RPro88 help to stabilize the interaction between UbcH5 and RING1b. | In the SPA motif of the UbcH5 (L7 residues USer94, UPro95, and UAla96), the sidechain hydroxyl of USer94 makes a hydrogen bond with the backbone carbonyl of RPro88 (from RING1b). Hydrophobic interactions between UPro95 and UAla96 with RIle53 and RPro88 help to stabilize the interaction between UbcH5 and RING1b. | ||
| - | |||
Canonical PRC1 such as BMI1/RING1b have intrinsically very low enzymatic activity compared with non-canonical PRC1, although there are only subtle differences between the structure of canonical and non-canonical complexes. Two charged helix alpha 3 residues present in a modeled BMI1 (ref taherbhoy 2015) seems responsible for the low activity of BMI1/RING1b, K73 and D77 form a salt bridge that may limit efficient ubiquitin transfer. In a computational modeling, BMI1 K73 clashes sterically with Ubiquitin, which implies that K73 must move to allow Ub to bind in an activated conformation or that Ub adopts a less optimal position during monoubiquitination. Either of these events could provide an energy barrier, slowing transfer catalyzed by BMI1. The intrinsically low activity of the BMI1/RING1b is offset by a relatively favorable interaction between E3–E2-Ub and nucleosome substrate, resulting in a site-specific monoubiquitination efficient enough. The energy barrier may be responsible for increasing the fidelity of the transfer to the appropriate substrate. Also, canonical and non-canonical differ in targeting sub-units, target genomic loci, and genes expression regulation. | Canonical PRC1 such as BMI1/RING1b have intrinsically very low enzymatic activity compared with non-canonical PRC1, although there are only subtle differences between the structure of canonical and non-canonical complexes. Two charged helix alpha 3 residues present in a modeled BMI1 (ref taherbhoy 2015) seems responsible for the low activity of BMI1/RING1b, K73 and D77 form a salt bridge that may limit efficient ubiquitin transfer. In a computational modeling, BMI1 K73 clashes sterically with Ubiquitin, which implies that K73 must move to allow Ub to bind in an activated conformation or that Ub adopts a less optimal position during monoubiquitination. Either of these events could provide an energy barrier, slowing transfer catalyzed by BMI1. The intrinsically low activity of the BMI1/RING1b is offset by a relatively favorable interaction between E3–E2-Ub and nucleosome substrate, resulting in a site-specific monoubiquitination efficient enough. The energy barrier may be responsible for increasing the fidelity of the transfer to the appropriate substrate. Also, canonical and non-canonical differ in targeting sub-units, target genomic loci, and genes expression regulation. | ||
(fig destacando os resíduos K73 and D77 de BMI1 no complexo PRC1) | (fig destacando os resíduos K73 and D77 de BMI1 no complexo PRC1) | ||
| - | The central domain of BMI1 forms an ubiquitin-like (UBL) domain, which is involved in protein-protein interactions, including interactions with the transcription factors E4F1, Zfp277 and the PLZF-RARA fusion protein. The best characterized binding partners of the UBL domain are the polyhomeotic proteins (PHC1, PHC2, PHC3) (ref Gray 2016). The UBL domain binds a short, 24 amino acid fragment, of PHC2 in a b-hairpin conformation. Also, UBL domain is involved in homo-oligomerization of BMI1. NMR and carbon detected NMR found that residues 30-51 are strongly conserved between PHC2, PHC1 and PHC3 suggesting that BMI1 interacts with the three members of the polyhomeotic family in a very similar manner and with similar affinities. Deletion of the corresponding motif abolished the interaction with BMI1. In the PHC2-BMI1 complex, PHC2 residues 33–47 adopt a beta-hairpin conformation in the complex. The PHC2-BMI1 interaction involves an antiparallel b-sheet formed between the beta-hairpin of PHC2 and the beta 2 strand of BMI1 UBL, | + | The central domain of BMI1 forms an <scene name='78/787701/5fr6_bmi1/1'>ubiquitin-like (UBL) domain</scene>, which is involved in protein-protein interactions, including interactions with the transcription factors E4F1, Zfp277 and the PLZF-RARA fusion protein. The best characterized binding partners of the UBL domain are the polyhomeotic proteins (PHC1, PHC2, PHC3) (ref Gray 2016). The UBL domain binds a short, 24 amino acid fragment, of PHC2 in a b-hairpin conformation. Also, UBL domain is involved in homo-oligomerization of BMI1. NMR and carbon detected NMR found that residues 30-51 are strongly conserved between PHC2, PHC1 and PHC3 suggesting that BMI1 interacts with the three members of the polyhomeotic family in a very similar manner and with similar affinities. Deletion of the corresponding motif abolished the interaction with BMI1. In the <scene name='78/787701/2na1/1'>PHC2-BMI1 complex</scene>, PHC2 residues 33–47 adopt a <scene name='78/787701/2na1_betahairpin_highlighted/1'>beta-hairpin conformation</scene> in the complex, in greenyellow. The PHC2-BMI1 interaction involves an <scene name='78/787701/2na1_bhairpin_b2_highlighted/1'>antiparallel b-sheet</scene> formed between the beta-hairpin of PHC2 and the beta 2 strand of BMI1 UBL, in magenta. The antiparallel b-sheet is stabilized by the hydrogen bonds between BMI1 Tyr163 and PHC2 Gly46. |
(fig destacando as pontes de hidrogenio entre BMI1 Tyr163 e PHC2 Gly46, da beta-hairpin da PHC2 e folha beta 2 da BMI1 UBL) | (fig destacando as pontes de hidrogenio entre BMI1 Tyr163 e PHC2 Gly46, da beta-hairpin da PHC2 e folha beta 2 da BMI1 UBL) | ||
| Line 48: | Line 48: | ||
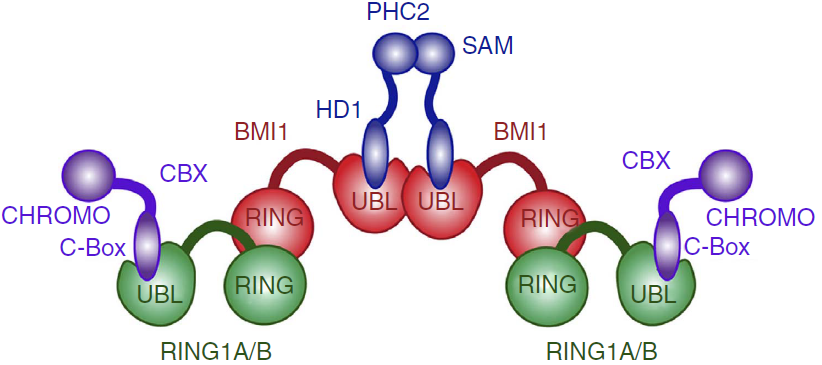
Besides biding to PHC1-3, the UBL domain also has a propensity of forming homo-oligomers in solution. This tendency is probably due to one hydrophobic region of UBL domain, which mutation at Ile212 impairs homo-oligomerization of the BMI1–PHC2 complex and the homo-oligomerization of UBL domain itself. This suggests that the same hydrofobic region might be responsible for the oligomerization of the BMI1/RING1b complex, such as the PRC1 complex tetramer found in vitro (ref Buchwald 2006). | Besides biding to PHC1-3, the UBL domain also has a propensity of forming homo-oligomers in solution. This tendency is probably due to one hydrophobic region of UBL domain, which mutation at Ile212 impairs homo-oligomerization of the BMI1–PHC2 complex and the homo-oligomerization of UBL domain itself. This suggests that the same hydrofobic region might be responsible for the oligomerization of the BMI1/RING1b complex, such as the PRC1 complex tetramer found in vitro (ref Buchwald 2006). | ||
| - | + | [[Image:proposed architecture of the PRC1 complex oligomer.png]] | |
| - | + | ||
| + | Proposed architecture of the PRC1 complex oligomer.<ref name="ncomms">DOI: 10.1038/ncomms13343</ref> | ||
== Conservation == | == Conservation == | ||
Revision as of 13:11, 17 June 2018
| |||||||||||
References
- ↑ 1.0 1.1 Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. 2011 Jul 19. The EMBO Journal (2011) 30, 3285–3297
- ↑ Gray F, Cho HJ, Shukla S, He S, Harris A, Boytsov B, Jaremko L, Jaremko M, Demeler B, Lawlor ER, Grembecka J, Cierpicki T. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat Commun. 2016 Nov 9;7:13343. doi: 10.1038/ncomms13343. PMID:27827373 doi:http://dx.doi.org/10.1038/ncomms13343
https://drive.google.com/drive/folders/1l195aNuY6joOd74GKKxa-XWTRMBv_uWF?usp=sharing