This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Ricardo Alberto Chiong Zevallos/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='350' side='right' caption='Structure of a Bmi1 protein' scene='78/787701/Bmi1_isolada_da_2h0d/ | + | <StructureSection load='' size='350' side='right' caption='Structure of a Bmi1 protein' scene='78/787701/Bmi1_isolada_da_2h0d/3'> |
| Line 8: | Line 8: | ||
Monoubiquitinated H2A is enriched on the inactive human female X-chromosome. The inactivation of one of the X-chromosome is responsible for “dosing” the X-chromosome expression, which leads to the physiological X expression levels in females similar to the X expression levels in males (which has only one X). | Monoubiquitinated H2A is enriched on the inactive human female X-chromosome. The inactivation of one of the X-chromosome is responsible for “dosing” the X-chromosome expression, which leads to the physiological X expression levels in females similar to the X expression levels in males (which has only one X). | ||
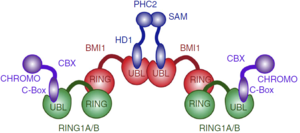
| - | PcG PRC1 is responsible for chromatin remodeling and histone modification, mediating monoubiquitination of histone H2A at Lysine 119, rendering chromatin heritably changed in its expressibility. BMI1 regulates the E3 ubiquitin-protein ligase activity of RING1b, which is also a protein component of the PcG PRC1 complex | + | PcG PRC1 is responsible for chromatin remodeling and histone modification, mediating monoubiquitination of histone H2A at Lysine 119, rendering chromatin heritably changed in its expressibility. BMI1 regulates the E3 ubiquitin-protein ligase activity of RING1b, which is also a protein component of the PcG PRC1 complex. |
| - | The PRC1 complex is composed of four core subunits: CBX (polycomb; CBX2/4/6/7/8), PCGF (polycomb group factors; PCGF1–6, one of which is the BMI1), PHC (polyhomeotic homologues; PHC1/2/3) and RING E3 ligase (RING1A/B). However, a complex formed just by a polycomb group factor and RING1b is sufficient for the E3 ubiquitin ligase activity, being BMI1 the PCGF4. The PCGF2/RING1b (being PCGF2 the MEL-18 protein) and BMI1/RING1b represent canonical PRC1 complex, while PCGF1, PCGF 3, PCGF5 and PCGF 6 forms non-canonical PRC1 complexes. | ||
| - | + | == Structure == | |
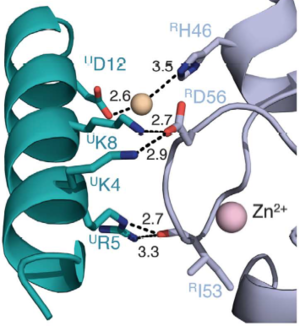
| + | BMI1 is a 37 kDa protein composed of three distinct regions: an N-terminal RING domain, a central domain and a C-terminal proline-serine rich domain involved in the regulation of protein stability <ref>DOI: 10.1038/ncomms13343</ref> . In vitro, BMI1 heterodimerize with RING1b via their N-terminal RING domains, forming an sufficient active E3 ubiquitin ligase. The heterodimerization occurs by the wrapping of the N-terminal arm of Ring1b around a groove on the surface of Bmi1, which stabilizes the structure of Ring1b and greatly stimulates ligase activity. Is worth noting that RING-domains, such as the found in BMI1 and RING1b, are structurally stabilized through the binding of two zinc atoms, hence the <scene name='78/787701/Zn_highlighted_bmi1-ring1b/2'>BMI1/RING1b complex</scene> interacts with 4 Zinc atoms to the correct folding (Zn highlighted in magenta). X-ray crystallography shows prominent salt bridges being formed by the Bmi1/Ring1b pairs of Asp72/Arg70, Glu11/Lys112, Lys81/Glu48, and Thr41/Arg26 <ref>doi: 10.1038/emboj.2011.243</ref>. However, in vivo, the PRC1 complex is composed of four core subunits: CBX (polycomb; CBX2/4/6/7/8), PCGF (polycomb group factors; PCGF1–6, being PCGF4 the BMI1), PHC (polyhomeotic homologues; PHC1/2/3) and RING E3 ligase (RING1A/B). The PCGF2/RING1b (being PCGF2 the MEL-18 protein) and BMI1/RING1b represent canonical PRC1 complex, while PCGF1, PCGF 3, PCGF5 and PCGF 6 form non-canonical PRC1 complexes when in pair with RING1a or RING1b. | ||
| - | + | In the complex, <scene name='78/787701/Bmi1_destacada_de_2h0d/2'>BMI1</scene> and <scene name='78/787701/Ring1b_destacada_de_2h0d/2'>RING1b</scene> form a heterodimer and only RING1b interacts with <scene name='78/787701/Ubiquitin-conjugating_enzyme_u/2'>UbcH5c</scene>, which is a Ubiquitin Conjugating Enzyme E2. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | . | + | |
A RING-domain E3 is responsible for promoting the transfer of ubiquitin (Ub) from the active site of the E2 ubiquitin-conjugating enzyme to an acceptor lysine residue in the substrate. The PRC1 complex promote the monoubiquitination of histone H2A on lysine 119 (uH2A), which leads to the stalling of the RNA polymerase at the promotor of the monoubiquitinated gene, hence the transcription doesn't occur. | A RING-domain E3 is responsible for promoting the transfer of ubiquitin (Ub) from the active site of the E2 ubiquitin-conjugating enzyme to an acceptor lysine residue in the substrate. The PRC1 complex promote the monoubiquitination of histone H2A on lysine 119 (uH2A), which leads to the stalling of the RNA polymerase at the promotor of the monoubiquitinated gene, hence the transcription doesn't occur. | ||
| Line 58: | Line 54: | ||
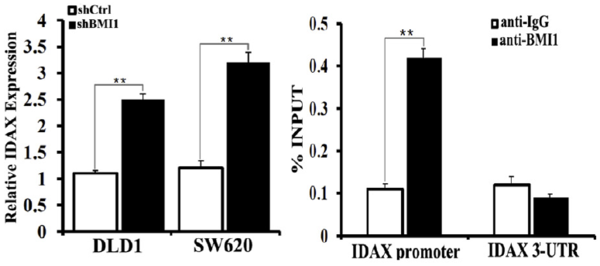
BMI1 is also associated with colon cancer, in which BMI1 activates WNT signaling mainly through the downregulation of IDAX expression. The BMI1 protein binds to the promoter of IDAX, a Wnt antagonist, and decreases it’s transcription. The WNT signaling pathway is very important in cell fate determination and tissue development and stem cell maintenance. Excessive activation of this pathway leads to tumorigenesis in several types of human cancers. According to The Cancer Genome Atlas Network, the WNT pathway is hyper-activated in over 90% of colon cancer. | BMI1 is also associated with colon cancer, in which BMI1 activates WNT signaling mainly through the downregulation of IDAX expression. The BMI1 protein binds to the promoter of IDAX, a Wnt antagonist, and decreases it’s transcription. The WNT signaling pathway is very important in cell fate determination and tissue development and stem cell maintenance. Excessive activation of this pathway leads to tumorigenesis in several types of human cancers. According to The Cancer Genome Atlas Network, the WNT pathway is hyper-activated in over 90% of colon cancer. | ||
| - | [[Image:BMI1 regulates IDAX transcription.png| | + | [[Image:BMI1 regulates IDAX transcription.png|600px]] |
IDAX as a target from BMI1. In the left is shown that the BMI1 knockdown by short hairpin RNAs leads to the increased IDAX transcription in colon cancer cell lines. In the right is shown the binding of the BMI1 protein to the promoter of IDAX, analyzed by ChIP-PCR. | IDAX as a target from BMI1. In the left is shown that the BMI1 knockdown by short hairpin RNAs leads to the increased IDAX transcription in colon cancer cell lines. In the right is shown the binding of the BMI1 protein to the promoter of IDAX, analyzed by ChIP-PCR. | ||
Revision as of 22:06, 17 June 2018
| |||||||||||
References
- ↑ Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999 Jan 14;397(6715):164-8. doi: 10.1038/16476. PMID:9923679 doi:http://dx.doi.org/10.1038/16476
- ↑ Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004 Oct 14;431(7010):873-8. Epub 2004 Sep 22. PMID:15386022 doi:10.1038/nature02985
- ↑ Gray F, Cho HJ, Shukla S, He S, Harris A, Boytsov B, Jaremko L, Jaremko M, Demeler B, Lawlor ER, Grembecka J, Cierpicki T. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat Commun. 2016 Nov 9;7:13343. doi: 10.1038/ncomms13343. PMID:27827373 doi:http://dx.doi.org/10.1038/ncomms13343
- ↑ Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011 Jul 19. doi: 10.1038/emboj.2011.243. PMID:21772249 doi:10.1038/emboj.2011.243
- ↑ Taherbhoy AM, Huang OW, Cochran AG. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat Commun. 2015 Jul 7;6:7621. doi: 10.1038/ncomms8621. PMID:26151332 doi:http://dx.doi.org/10.1038/ncomms8621
External resources
- Aggregated information about BMI1 protein in UniProt database
- Aggregated information about BMI1 gene in GeneCards database
https://drive.google.com/drive/folders/1l195aNuY6joOd74GKKxa-XWTRMBv_uWF?usp=sharing