Kisker lab: 5B5Q
From Proteopedia
(→Structure) |
(→Structure) |
||
| Line 18: | Line 18: | ||
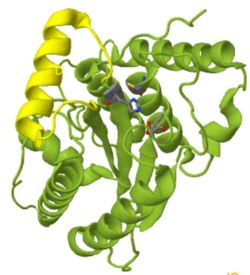
<scene name='78/781027/Panela/3'>Panel A:</scene> The overall structure, shown as cartoon, has a fold common to other deubiquitinases (green) with a helix inserted between strand 1 and 2 (yellow). The protein belongs to the family of cysteine proteases, in which an active-site cysteine initiates hydrolysis by acting as a nucleophile. Just like serine proteases, cystein proteases have a catalytic triad (i.e. three highly conserved residues in the active site). The catalytic triad is shown in all-bonds representation. | <scene name='78/781027/Panela/3'>Panel A:</scene> The overall structure, shown as cartoon, has a fold common to other deubiquitinases (green) with a helix inserted between strand 1 and 2 (yellow). The protein belongs to the family of cysteine proteases, in which an active-site cysteine initiates hydrolysis by acting as a nucleophile. Just like serine proteases, cystein proteases have a catalytic triad (i.e. three highly conserved residues in the active site). The catalytic triad is shown in all-bonds representation. | ||
| - | (If you are new to proteopedia: Click on the green links in the text to see the scene in the 3D browser on the right. Use the mouse to change the view of the scene. Click on the controls on the bottom of the 3D browser to automatically spin the molecule, to resize the 3D browser window | + | (If you are new to proteopedia: Click on the green links in the text to see the scene in the 3D browser on the right. Use the mouse to change the view of the scene. Click on the controls on the bottom of the 3D browser to automatically spin the molecule, to make the rendering fancier (and perhaps slower), to resize the 3D browser window or to open a pop-up window with the same content. More controls become visible when you right-click inside the 3D browser area, and you can even remove parts of the scene or add to it using these controls.) |
===Related proteins=== | ===Related proteins=== | ||
Revision as of 00:13, 26 June 2018
Contents |
How this page was created
The goal of this page is to provide three-dimensional and interactive figures to explore the structure of a protein involved in bacterial infections (PDB code 5B5Q). The starting point are the figures found in the paper describing the 5B5Q structure. Biochemistry students from Westfield State University recreated these figures as best as possible in jmol, and revised them after getting feedback from researchers who authored the primary citation (Kisker lab in Würzburg, Germany). A special thank you goes to Ose Aimua, Nina Aldabayeva, Faiqa Ashraf, Kaleigh Florek, Ellie Hoeg, Aya Maytham, Christian Mikule, Brigid Murray, Kevin Pelletier, Brandon Reder, Erin Riley, Brian Schuler, and Jakob Wyman for the revisions of figures and for working on the links to other proteopedia pages.
If you are interested to see the jmol scripts used to make these figures, take a look at the discussion page (2nd tab above).
Chlamydia trachomatis inhibits apoptosis
Chlamydia trachomatis is a bacterium that reproduces inside human cells. One defense of the human body against Chlamydia is to kill affected cells before Chlamydia reproduces. This is done through a process called apoptosis, programmed cell death. One player in apoptosis is the human protein Mcl-1. High Mcl-1 levels inhibit one of the signalling pathways that lead to apoptosis. Chlamydia inhibits Mcl-1 degradation so that Mcl-1 levels remain high.
Protein ubiquitination and degradation
Human cells have a protein assembly called the proteasome, which specializes in degrading proteins. Ubiquitin is a small, highly soluble protein; when ubiquitin chains are attached to other proteins in a certain way, it acts as a signal for protein degradation. The proteasome only degrades proteins that are poly-ubiquitinated, i.e. are covalently linked to a linear chain of ubiquitins. The covalent link is between the amino group of a lysine side chain and the carboxylic acid of a glycine at the C-terminus of ubiquitin.
The deubiquitinase activity of Cdu1 stabilizes Mcl-1
The Chlamydia protein Cdu1 is a protease that catalyzes the hydrolysis of ubiquitin chains from Mcl-1. When polyubiquitinated, Mcl-1 is destined to be degraded by the proteasome, lowering the level of Mcl-1 and subsequently leading to apoptosis. The activity of Cdu1 counteracts this by removing the ubiquitin, thus leading to higher levels of Mcl-1 in the cell.
Structure
| |||||||||||
Relevant Links
5b5q : autogenerated Proteopedia page on coordinates
Ubiquitin Structure & Function : Proteopedia article on Ubiquitin Structure, function and conjugation
Ubiquitin chains : Proteopedia article on Ubiquitin Chains
Category: Kisker C : list of structures associated with the Kisker lab