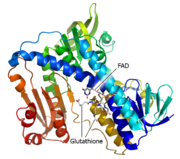
Glutathione Reductase
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
=Reaction= | =Reaction= | ||
| - | [[Image:Glutathione _reductase_mechanism.gif|left | + | [[Image:Glutathione _reductase_mechanism.gif|left|thumb|Reaction]] |
{{Clear}} | {{Clear}} | ||
The action of glutathione reductase proceeds through a cyclic series of structures in differing redox states<ref name="main"/>. NADPH binds causing a transient reduction of flavin and this reduced flavin consequently reduces Cys58-Cys63 disulfide bond, forming a short lived covalent intermediate with Cys63<ref name="main"/>. Following this, a stable charge-transfer complex between flavin and the Cys63 thiolate forms<ref name="main"/>. After formation the NADP+ dissociates and is replaced by another NADPH<ref name="main"/>. This is the end of the reductive first half of the mechanism and the oxidative half is initiated upon the binding of GSSG<ref name="main"/>. The Cys58 in glutathione reductase attacks CysI of the GSSG to form a mixed disulfide between the first GS and Cys58<ref name="main"/>. The second GSH is the free to leave and the disulfide bond is reformed between Cys58 and Cys63 of glutathione reductase. Finally, the first molecule of GSH is released<ref name="main"/>. The glutathione reductase is then able to be recycled to allow for the binding of NADPH once again<ref name="main"/>. | The action of glutathione reductase proceeds through a cyclic series of structures in differing redox states<ref name="main"/>. NADPH binds causing a transient reduction of flavin and this reduced flavin consequently reduces Cys58-Cys63 disulfide bond, forming a short lived covalent intermediate with Cys63<ref name="main"/>. Following this, a stable charge-transfer complex between flavin and the Cys63 thiolate forms<ref name="main"/>. After formation the NADP+ dissociates and is replaced by another NADPH<ref name="main"/>. This is the end of the reductive first half of the mechanism and the oxidative half is initiated upon the binding of GSSG<ref name="main"/>. The Cys58 in glutathione reductase attacks CysI of the GSSG to form a mixed disulfide between the first GS and Cys58<ref name="main"/>. The second GSH is the free to leave and the disulfide bond is reformed between Cys58 and Cys63 of glutathione reductase. Finally, the first molecule of GSH is released<ref name="main"/>. The glutathione reductase is then able to be recycled to allow for the binding of NADPH once again<ref name="main"/>. | ||
Revision as of 14:26, 5 July 2018
Glutathione reductase, also known as GSH reductase, is found in the human cells and converts oxidized glutathione (GSSG) to two molecules of reduced gluthatione (GSH) [1]. EC:1.8.1.7. For more details see Molecular Playground/Glutathione Reductase.
| |||||||||||
3D structures of glutathione reductase
Updated on 05-July-2018
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 Berkholz DS, Faber HR, Savvides SN, Karplus PA. Catalytic cycle of human glutathione reductase near 1 A resolution. J Mol Biol. 2008 Oct 3;382(2):371-84. Epub 2008 Jul 7. PMID:18638483 doi:10.1016/j.jmb.2008.06.083
- ↑ Holleschau AM, Rathbun WB. Thermal inactivation study of glutathione peroxidase and glutathione reductase activities in lenses of primates and non-primates. Curr Eye Res. 1991 Mar;10(3):221-9. PMID:2044390
- ↑ 3.0 3.1 Kelner MJ, Montoya MA. Structural organization of the human glutathione reductase gene: determination of correct cDNA sequence and identification of a mitochondrial leader sequence. Biochem Biophys Res Commun. 2000 Mar 16;269(2):366-8. PMID:10708558 doi:10.1006/bbrc.2000.2267
- ↑ 4.0 4.1 Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001 Sep;10(9):1712-28. PMID:11514662 doi:10.1110/ps.12801
- ↑ 5.0 5.1 Masella R, Di Benedetto R, Vari R, Filesi C, Giovannini C. Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes. J Nutr Biochem. 2005 Oct;16(10):577-86. PMID:16111877 doi:10.1016/j.jnutbio.2005.05.013
- ↑ Pastore A, Federici G, Bertini E, Piemonte F. Analysis of glutathione: implication in redox and detoxification. Clin Chim Acta. 2003 Jul 1;333(1):19-39. PMID:12809732
- ↑ Masella R, Di Benedetto R, Vari R, Filesi C, Giovannini C. Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes. J Nutr Biochem. 2005 Oct;16(10):577-86. PMID:16111877 doi:10.1016/j.jnutbio.2005.05.013
- ↑ Voehringer DW, Hirschberg DL, Xiao J, Lu Q, Roederer M, Lock CB, Herzenberg LA, Steinman L, Herzenberg LA. Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2680-5. PMID:10716996
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 Kamerbeek NM, van Zwieten R, de Boer M, Morren G, Vuil H, Bannink N, Lincke C, Dolman KM, Becker K, Schirmer RH, Gromer S, Roos D. Molecular basis of glutathione reductase deficiency in human blood cells. Blood. 2007 Apr 15;109(8):3560-6. Epub 2006 Dec 21. PMID:17185460 doi:10.1182/blood-2006-08-042531