Malate dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 66: | Line 66: | ||
**[[4i1i]] – LmMd + NAD – ''Leishmania major''<br /> | **[[4i1i]] – LmMd + NAD – ''Leishmania major''<br /> | ||
**[[5ujk]] – MeMd + NAD - ''Methylobacterium extorquens'' <br /> | **[[5ujk]] – MeMd + NAD - ''Methylobacterium extorquens'' <br /> | ||
| + | **[[5nuf]], [[5nue]] – Md1 + NAD – ''Arabidosis thaliana''<br /> | ||
**[[4uup]] – Md + NADH - ''Trichiminad'' <br /> | **[[4uup]] – Md + NADH - ''Trichiminad'' <br /> | ||
**[[5kvv]] – MtMd + NADH – ''Mycobacterium tuberculosis'' <br /> | **[[5kvv]] – MtMd + NADH – ''Mycobacterium tuberculosis'' <br /> | ||
| + | **[[6bal]] – HiMd + malate - Haemophilus influenzae<br /> | ||
*Md ternary complex | *Md ternary complex | ||
| Line 105: | Line 107: | ||
**[[4ror]] – MeMd <br /> | **[[4ror]] – MeMd <br /> | ||
**[[4tvo]] – Md – ''Mycobacterium tubrtculosis'' <br /> | **[[4tvo]] – Md – ''Mycobacterium tubrtculosis'' <br /> | ||
| + | **[[6aoo]] – HiMd <br /> | ||
| + | **[[5ulv]] – MeMd <br /> | ||
| + | **[[5nfr]] – Md – ''Plasmodium falciparum''<br /> | ||
| + | **[[4uuo]] – Md – ''Trichomonas vaginalis''<br /> | ||
}} | }} | ||
Revision as of 09:13, 2 August 2018
| |||||||||||
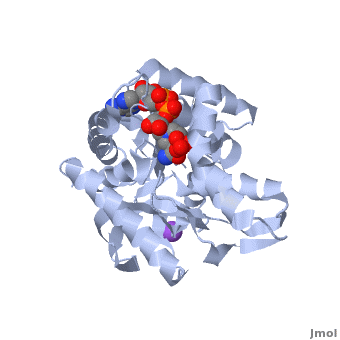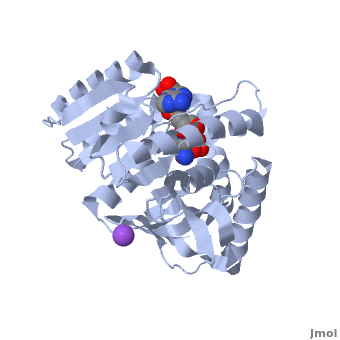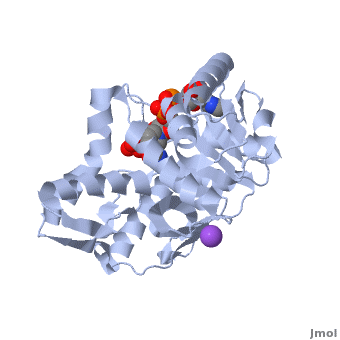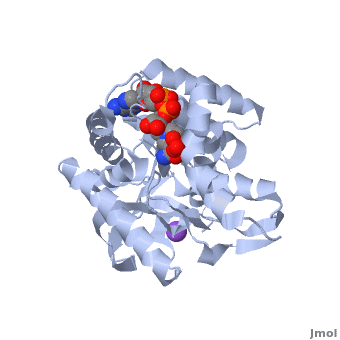
3D Structures of Malate Dehydrogenase
Updated on 02-August-2018
The holo-MDH contains NAD or its derivatives while the apo-MDH lacks it.
Additional Resources
References
- ↑ Minarik P, Tomaskova N, Kollarova M, Antalik M. Malate dehydrogenases--structure and function. Gen Physiol Biophys. 2002 Sep;21(3):257-65. PMID:12537350
- ↑ Matsuda T, Takahashi-Yanaga F, Yoshihara T, Maenaka K, Watanabe Y, Miwa Y, Morimoto S, Kubohara Y, Hirata M, Sasaguri T. Dictyostelium Differentiation-Inducing Factor-1 Binds to Mitochondrial Malate Dehydrogenase and Inhibits Its Activity. J Pharmacol Sci. 2010 Feb 20. PMID:20173310
- ↑ Matsuda T, Takahashi-Yanaga F, Yoshihara T, Maenaka K, Watanabe Y, Miwa Y, Morimoto S, Kubohara Y, Hirata M, Sasaguri T. Dictyostelium Differentiation-Inducing Factor-1 Binds to Mitochondrial Malate Dehydrogenase and Inhibits Its Activity. J Pharmacol Sci. 2010 Feb 20. PMID:20173310
- ↑ Musrati RA, Kollarova M, Mernik N, Mikulasova D. Malate dehydrogenase: distribution, function and properties. Gen Physiol Biophys. 1998 Sep;17(3):193-210. PMID:9834842
- ↑ Boernke WE, Millard CS, Stevens PW, Kakar SN, Stevens FJ, Donnelly MI. Stringency of substrate specificity of Escherichia coli malate dehydrogenase. Arch Biochem Biophys. 1995 Sep 10;322(1):43-52. PMID:7574693 doi:http://dx.doi.org/10.1006/abbi.1995.1434
- ↑ Plancarte A, Nava G, Mendoza-Hernandez G. Purification, properties, and kinetic studies of cytoplasmic malate dehydrogenase from Taenia solium cysticerci. Parasitol Res. 2009 Jul;105(1):175-83. Epub 2009 Mar 10. PMID:19277715 doi:10.1007/s00436-009-1380-6
- ↑ Goward CR, Nicholls DJ. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 1994 Oct;3(10):1883-8. PMID:7849603 doi:http://dx.doi.org/10.1002/pro.5560031027
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Jake Ezell, Joel L. Sussman, Joshua Johnson, Angel Herraez, Jaime Prilusky, David Canner