We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Phillips Academy Computer-Aided Protein Visualization Lab
From Proteopedia
(Difference between revisions)
(New page: == Introduction to Computer-Aided Protein Visualization Lab == <StructureSection load='1pgb' size='340' side='right' caption='This simple protein, B1 Immunoglobulin-binding domain of Strep...) |
|||
| Line 5: | Line 5: | ||
One particularly key area of current research is the design of drugs against specific protein targets. Scientists may look for drugs to block the activity of an enzyme of an attacking bacteria, protist or virus. After first finding an enzyme that is slightly different between humans and the invading species, the scientists can then use computers to look at the enzyme and try to fit tens of thousands of compounds into the enzyme in such a way as to block its activity; most commonly this involves plugging the active site of the invading species enzyme selectively. Using computers to analyze this problem can speed up the screening of 50,000 potential drugs from many years down to one week! Once a few compounds with potential are approved by the computer, the scientist can look to chemically modify those compounds to make them even better and then try them out in drug trials against the enzyme in test tubes, and eventually in drug trials in animals and humans. | One particularly key area of current research is the design of drugs against specific protein targets. Scientists may look for drugs to block the activity of an enzyme of an attacking bacteria, protist or virus. After first finding an enzyme that is slightly different between humans and the invading species, the scientists can then use computers to look at the enzyme and try to fit tens of thousands of compounds into the enzyme in such a way as to block its activity; most commonly this involves plugging the active site of the invading species enzyme selectively. Using computers to analyze this problem can speed up the screening of 50,000 potential drugs from many years down to one week! Once a few compounds with potential are approved by the computer, the scientist can look to chemically modify those compounds to make them even better and then try them out in drug trials against the enzyme in test tubes, and eventually in drug trials in animals and humans. | ||
| - | == First some background: (make sure that you understand the | + | == First some background: (make sure that you understand the '''bold''' words)== |
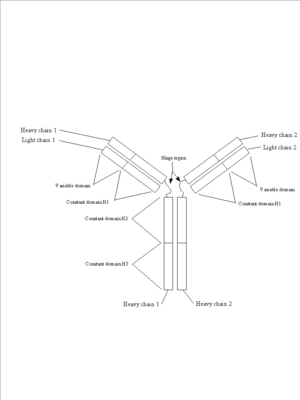
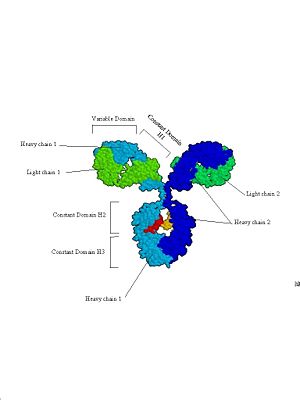
| - | Proteins are synthesized on ribosomes by linking together many amino acids into a long chain. If you could observe a protein as it is made, it would look like a string of pearls (amino acids) feeding out the end of the ribosome as it floats in the cytoplasm of the cell ([https://www.dnalc.org/view/15501-Translation-RNA-to-protein-3D-animation-with-basic-narration.html Video of Translation (DNALC)]). This structure is called the primary structure (or 1° structure) and refers to the sequence of amino acids of the protein. After protein synthesis has started, | + | '''Proteins''' are synthesized on '''ribosomes''' by linking together many '''amino acids''' into a long chain. If you could observe a protein as it is made, it would look like a string of pearls (amino acids) feeding out the end of the ribosome as it floats in the '''cytoplasm''' of the '''cell''' ([https://www.dnalc.org/view/15501-Translation-RNA-to-protein-3D-animation-with-basic-narration.html Video of Translation (DNALC)]). This structure is called the '''primary structure''' (or 1° structure) and refers to the sequence of amino acids of the protein. After protein synthesis has started, the sequence of amino acids will begin to fold into a 3-dimensional structure. This process is called '''protein folding''': |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Protein folding: | Protein folding: | ||
| - | The most important rule about protein structure is that it is determined by the primary sequence of the protein. Protein folding is a complicated multi-step process. The first step results in the secondary structure (or 2o structure) of the protein. Secondary structures come in two flavors: alpha helices and beta sheets (or beta-pleated sheets). Alpha helices are spiral staircase structures (see structure 1 below), and beta-pleated sheets are flat regions where the amino acids run back and forth next to each other in long ribbons (see structure 2 below). These two structures form spontaneously based on the shape/hydrophobicity/charges of the amino acids and are held together by hydrogen bonds. The protein will now look like a string of pearls with twists or zig-zags at intervals along its length. | + | The most important rule about protein structure is that it is determined by the primary sequence of the protein. Protein folding is a complicated multi-step process. The first step results in the '''secondary structure''' (or 2o structure) of the protein. Secondary structures come in two flavors: '''alpha helices''' and '''beta sheets''' (or beta-pleated sheets). Alpha helices are spiral staircase structures (see structure 1 below), and beta-pleated sheets are flat regions where the amino acids run back and forth next to each other in long ribbons (see structure 2 below). These two structures form spontaneously based on the shape/'''hydrophobicity'''/'''charges''' of the amino acids and are held together by '''hydrogen bonds'''. The protein will now look like a string of pearls with twists or zig-zags at intervals along its length. |
1. <scene name='71/713432/Protein_secondary_structure/3'>Click to see alpha helix</scene> | 1. <scene name='71/713432/Protein_secondary_structure/3'>Click to see alpha helix</scene> | ||
| Line 21: | Line 17: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | The second step of protein folding results in the tertiary structure (or 3° structure). Tertiary structure gives the protein an overall three-dimensional structure. The tertiary structure of a protein is determined by a combination of factors including hydrogen bonds, ionic bonds (between positively and negatively charged amino acids), covalent disulfide bonds (between cysteine residues), and Van der Waals interactions. Tertiary structure can also be affected by repulsive forces between similarly charged amino acids, as well as hydrophobic and hydrophilic interactions with a solvent (commonly water). At a distance many proteins form what look to be large globs at this point, and it is only upon more careful and close up inspection that one can see the true uniqueness of the shape. | + | The second step of protein folding results in the '''tertiary structure''' (or 3° structure). Tertiary structure gives the protein an overall three-dimensional structure. The tertiary structure of a protein is determined by a combination of factors including hydrogen bonds, '''ionic bonds''' (between positively and negatively charged amino acids), '''covalent''' '''disulfide bonds''' (between cysteine residues), and '''Van der Waals''' interactions. Tertiary structure can also be affected by repulsive forces between similarly charged amino acids, as well as '''hydrophobic''' and '''hydrophilic''' interactions with a solvent (commonly water). At a distance many proteins form what look to be large globs at this point, and it is only upon more careful and close up inspection that one can see the true uniqueness of the shape. |
<br> | <br> | ||
[[Image:Intramolecular forces in tertiary structures.png]] | [[Image:Intramolecular forces in tertiary structures.png]] | ||
Revision as of 02:06, 3 September 2018
Introduction to Computer-Aided Protein Visualization Lab
| |||||||||||