We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Phage integrase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
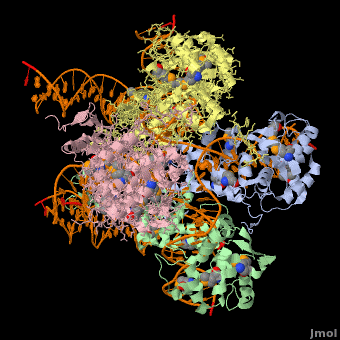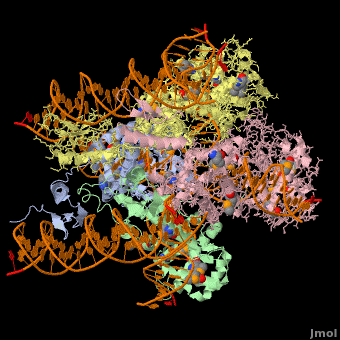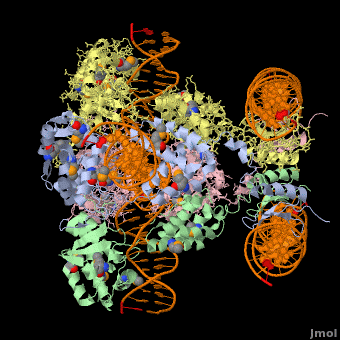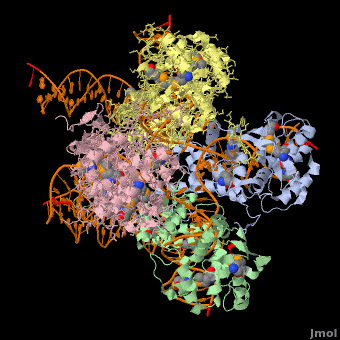
*The biological assembly of Enterobacteria phage λ integrase is <scene name='44/443479/Cv/3'>homotetramer</scene>. | *The biological assembly of Enterobacteria phage λ integrase is <scene name='44/443479/Cv/3'>homotetramer</scene>. | ||
</StructureSection> | </StructureSection> | ||
| - | == 3D Structures of phage integrase == | + | == 3D Structures of phage integrase see [[Retroviral Integrase]]== |
| - | ''Update June 2011'' | ||
| - | |||
| - | [[3nrw]] – PIN/site-specific recombinase N-terminal – ''Haloarcula marismortui''<br /> | ||
| - | [[3jtz]] – PIN arm-type binding domain – ''Yersinia pestis''<br /> | ||
| - | [[3ju0]] – HPI PIN arm-type binding domain – ''Pectobacterium atrosepticum''<br /> | ||
| - | [[2kkp]] – PIN SAM-like domain – ''Moorella thermoacetica'' – NMR<br /> | ||
| - | [[2kkv]] – PIN fragment – ''Salmonella enterica'' – NMR<br /> | ||
| - | [[2khq]] - PIN fragment – ''Staphylococcus saprophyticus'' – NMR<br /> | ||
| - | [[2wcc]] – λPIN DBD+ DNA – Enterobacteria phage λ - NMR<br /> | ||
| - | [[1kjk]] – λPIN N-terminal - NMR<br /> | ||
| - | [[2oxo]], [[1z19]], [[1z1b]] - λPIN core binding domain + DNA<br /> | ||
| - | [[1ae9]] - λPIN catalytic core domain (mutant) <br /> | ||
| - | [[1z1g]] – λPIN + DNA (Holliday junction)<br /> | ||
| - | [[1p7d]] – λPIN fragment + DNA <br /> | ||
| - | [[3bvp]] – PIN catalytic core domain – Lactococcus phage TP101-1<br /> | ||
| - | [[1aih]] - PIN catalytic core domain – Haemophilus phage HP1<br /> | ||
| - | [[2khv]], [[2kj5]] – PIN residues 102-199 – ''Nitrosospira multiformis'' - NMR | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D Structures of phage integrase see Retroviral Integrase
References
- ↑ Groth AC, Calos MP. Phage integrases: biology and applications. J Mol Biol. 2004 Jan 16;335(3):667-78. PMID:14687564 doi:http://dx.doi.org/10.1016/S0022283603013561