We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
RuBisCO
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
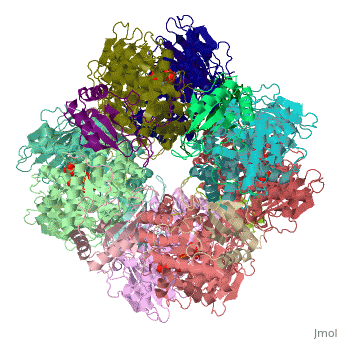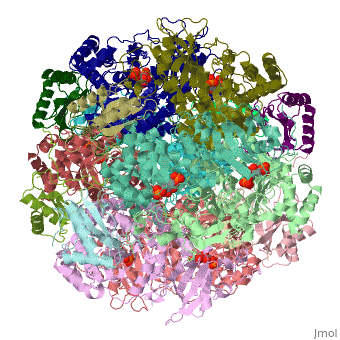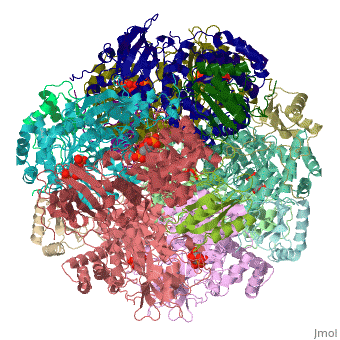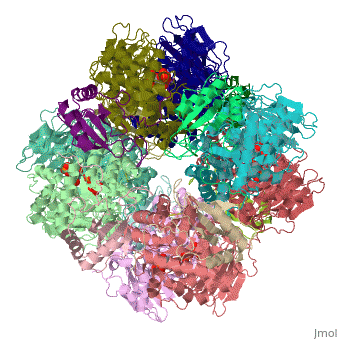
| - | <StructureSection load='1rcx' size=' | + | <StructureSection load='1rcx' size='350' side='right' caption='Spinach RuBisCO 8 large and 8 small chains complex with substrate ribulose-1,5- bisphosphate, [[1rcx]]'> |
== Function == | == Function == | ||
| Line 22: | Line 22: | ||
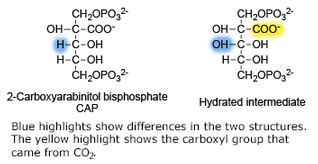
<scene name='46/463261/8ruc_active-site/10'>Residues that are involved in catalysis</scene> are shown shown here in CPK ball & stick. Asp 203 and Glu 204 bind to and position the magnesium ion. The carbamylated lysine residue KCX 201 coordinates Mg<sup>2+</sup> and initiates catalysis by extracting a proton from C3 of RUBP. Note the proximity of the carbamyl group to carbon 3 in this structure. His 294 acts as a catalytic base in the carboxylation step of the mechanism and accepts a proton from the hydroxyl of carbon 3. Mg<sup>2+</sup> is coordinated by six ligands. In addition to oxygen atoms in the three residues already mentioned, the ion binds to two oxygen atoms of RUBP. The 6th ligand is either water or in the carboxylation step it binds the incoming CO<sub>2</sub>. In the structure shown, Mg<sup>2+</sup> is bound to the carboxyl group in CAP that corresponds to the fixed CO<sub>2</sub> in the hydrated intermediate.</StructureSection> | <scene name='46/463261/8ruc_active-site/10'>Residues that are involved in catalysis</scene> are shown shown here in CPK ball & stick. Asp 203 and Glu 204 bind to and position the magnesium ion. The carbamylated lysine residue KCX 201 coordinates Mg<sup>2+</sup> and initiates catalysis by extracting a proton from C3 of RUBP. Note the proximity of the carbamyl group to carbon 3 in this structure. His 294 acts as a catalytic base in the carboxylation step of the mechanism and accepts a proton from the hydroxyl of carbon 3. Mg<sup>2+</sup> is coordinated by six ligands. In addition to oxygen atoms in the three residues already mentioned, the ion binds to two oxygen atoms of RUBP. The 6th ligand is either water or in the carboxylation step it binds the incoming CO<sub>2</sub>. In the structure shown, Mg<sup>2+</sup> is bound to the carboxyl group in CAP that corresponds to the fixed CO<sub>2</sub> in the hydrated intermediate.</StructureSection> | ||
| - | |||
| - | |||
| - | </StructureSection> | ||
==3D structures of RuBisCO== | ==3D structures of RuBisCO== | ||
| Line 34: | Line 31: | ||
** [[3rg6]], [[1rbl]] – SeRBCO – ''Synechococcus elongatus''<br /> | ** [[3rg6]], [[1rbl]] – SeRBCO – ''Synechococcus elongatus''<br /> | ||
** [[2ybv]] - RBCO – ''Thermosynechococcus elongatus''<br /> | ** [[2ybv]] - RBCO – ''Thermosynechococcus elongatus''<br /> | ||
| - | **[[4lf2]] – RpRBCO – ''Rhodopseudomonas palustris'' | + | **[[4lf2]], [[3qfw]] – RpRBCO – ''Rhodopseudomonas palustris''<br /> |
| - | + | ||
** [[1uzh]], [[1gk8]] – CrRBCO – ''Chlamydomonas reinhardtii''<br /> | ** [[1uzh]], [[1gk8]] – CrRBCO – ''Chlamydomonas reinhardtii''<br /> | ||
** [[1uw9]], [[1uwa]] – CrRBCO (mutant) <br /> | ** [[1uw9]], [[1uwa]] – CrRBCO (mutant) <br /> | ||
| Line 55: | Line 51: | ||
** [[4hhh]] – RBCO - pea<br /> | ** [[4hhh]] – RBCO - pea<br /> | ||
** [[5c2c]] - GaRBCO – ''Gallionella''<br /> | ** [[5c2c]] - GaRBCO – ''Gallionella''<br /> | ||
| + | ** [[5wsk]] – RBCO - wheat<br /> | ||
* RuBisCO complex with inhibitor 2-CABP | * RuBisCO complex with inhibitor 2-CABP | ||
| Line 74: | Line 71: | ||
** [[5iu0]] - RBCO + 2-CABP - ''Arabidopsis thaliana''<br /> | ** [[5iu0]] - RBCO + 2-CABP - ''Arabidopsis thaliana''<br /> | ||
** [[5mac]] - RBCO + 2-CABP - ''Methanococcoides burtonii''<br /> | ** [[5mac]] - RBCO + 2-CABP - ''Methanococcoides burtonii''<br /> | ||
| + | ** [[6ftl]] - RBCO + 2-CABP - ''Skeletonema marinoi''<br /> | ||
* RuBisCO complex with product | * RuBisCO complex with product | ||
Revision as of 21:46, 14 September 2018
| |||||||||||
3D structures of RuBisCO
Updated on 14-September-2018
See Also
References
- ↑ Andersson I. Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. J Mol Biol. 1996 May 31;259(1):160-74. PMID:8648644 doi:10.1006/jmbi.1996.0310
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alice Harmon, Joel L. Sussman, Alexander Berchansky